













| General Information: |
|
| Name(s) found: |
PSK2 /
YOL045W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1101 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
hexose metabolic process
[IGI protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein serine/threonine kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTYPVSAAAP ADISYSKNTP LVGLSKPPCL YQHASSSVDS FSSTFSDDDR SDLVAVPNES 60
61 PHAFSYNPIS PNSLGVRLTI LRRSLEIMVN SPDILHELKK KAPVIAYPPS LRHTRNLTET 120
121 ATLSASRDPL NGSLISPLVS NMPSPASRPV IQRATSLMVL PDNDTASKLN PAKSELENLL 180
181 FLLNLALENN SFERASDLHM LSLLNIKKIN FDSDIQKSET LKKVLLDSLA EPFFENYKKF 240
241 PHKDLGSKSQ YNEYEEKHDD IVSLADIKPQ QDYSRILHPF TSAKNSGPEA IFTCSQQYPW 300
301 NFKAANDLAC LTFGISKNVI KALTLLDLIH TDSRNFVLEK IMNAEDDNQE IVFTGETIPI 360
361 VQPNSTSNNN VPNLIWASLW AKRKNGLLVC VFEKTPCDYI DVMLNLRDFS VDSIIDTTHF 420
421 LENFDKKKQQ ESTSPMTEKK TVKFANEIHD IGSVSHSLSK LIDDVRFGKV FSADDDLLPL 480
481 SIRVANHVNE ERYFTLNCLS ENIPCAVTTS VLENEIKLKI HSLPYQAGLF IVDSHTLSLL 540
541 SFNKSVAKNM FGLRLHELAG SSVTKLVPSL ADMISYINKT YPMLNITLPE NKGLVLTEHF 600
601 FRKIEAEMHH DKDSFYTSIG LDGCHKDGNL IKVDVQLRVL NTNAVLLWIT HSRDVVIENY 660
661 TTVPSQLPML KENEIDVVGS RGSSSASSKK SSEKIPVNTL KAMADLSISS AETISNSDDE 720
721 VDLNQVNEKL RETSCGKVRG IESNDNNNYD DDMTMVDDPE LKHKIELTKM YTQDKSKFVK 780
781 DDNFKVDEKF IMRIIEPING EEIKKETNEL DKRNSTLKAT YLTTPEANIG SQKRIKKFSD 840
841 FTILQVMGEG AYGKVNLCIH NREHYIVVIK MIFKERILVD TWVRDRKLGT IPSEIQIMAT 900
901 LNKNSQENIL KLLDFFEDDD YYYIETPVHG ETGSIDLFDV IEFKKDMVEH EAKLVFKQVV 960
961 ASIKHLHDQG IVHRDIKDEN VIVDSHGFVK LIDFGSAAYI KSGPFDVFVG TMDYAAPEVL1020
1021 GGSSYKGKPQ DIWALGVLLY TIIYKENPYY NIDEILEGEL RFDKSEHVSE ECISLIKRIL1080
1081 TREVDKRPTI DEIYEDKWLK I |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PSK1 PSK2 |
| View Details | Krogan NJ, et al. (2006) |
|
AEP3 CDC20 FAB1 FOL2 GZF3 LSP1 NOP13 PIL1 PKH1 PRP16 PSK2 RGC1 RPL9B YLR179C YMR086W YTA6 |
| View Details | Ho Y, et al. (2002) |
|
FUN30 GPH1 HPR1 PSK1 PSK2 RPN1 RVB1 SET1 SFP1 SGS1 SUV3 SWD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
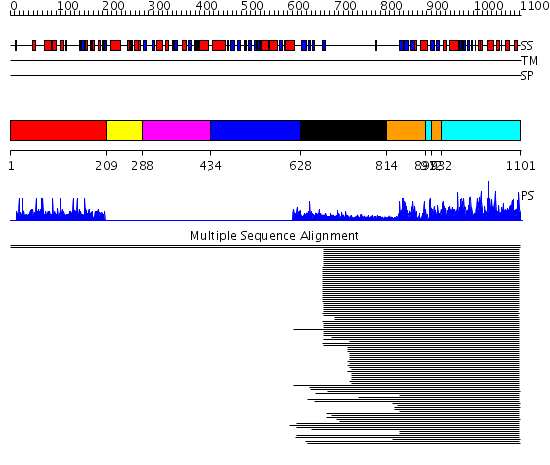
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | 1.002935 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [209..287] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [288..433] | 12.82 | N-terminal PAS domain of Pas kinase | |
| 4 | View Details | [434..627] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [628..813] | 23.0 | C2 domain from protein kinase c (alpha) | |
| 6 | View Details | [814..896] [912..931] |
318.228787 | cAMP-dependent PK, catalytic subunit | |
| 7 | View Details | [897..911] [932..1101] |
318.228787 | cAMP-dependent PK, catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)