













| General Information: |
|
| Name(s) found: |
FUN30 /
YAL019W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1131 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA |
| Biological Process: |
chromosome organization
[IGI]
chromatin silencing at rDNA [IMP] chromatin silencing at silent mating-type cassette [IMP][IPI] chromatin silencing at telomere [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGSHSNDED DVVQVPETSS PTKVASSSPL KPTSPTVPDA SVASLRSRFT FKPSDPSEGA 60
61 HTSKPLPSGS PEVALVNLAR EFPDFSQTLV QAVFKSNSFN LQSARERLTR LRQQRQNWTW 120
121 NKNASPKKSE TPPPVKKSLP LANTGRLSSI HGNINNKSSK ITVAKQKTSI FDRYSNVINQ 180
181 KQYTFELPTN LNIDSEALSK LPVNYNKKRR LVRADQHPIG KSYESSATQL GSAREKLLAN 240
241 RKYGRHANDN DEEEEESMMT DDDDASGDDY TESTPQINLD EQVLQFINDS DIVDLSDLSD 300
301 TTMHKAQLIA SHRPYSSLNA FVNTNFNDKD TEENASNKRK RRAAASANES ERLLDKITQS 360
361 IRGYNAIESV IKKCSSYGDL VTSQMKKWGV QVEGDNSELD LMNLGEDDDD DNDDGNNDNN 420
421 NSNNNNTAGA DATSKEKEDT KAVVEGFDET SAEPTPAPAP APVERETKRI RNTTKPKVVE 480
481 DEDDDVDLEA IDDELPQSEH EDDDYEEEDE DYNDEEEDVE YDDGDDDDDD DDEFVATRKN 540
541 THVISTTSRN GRKPIVKFFK GKPRLLSPEI SLKDYQQTGI NWLNLLYQNK MSCILADDMG 600
601 LGKTCQVISF FAYLKQINEP GPHLVVVPSS TLENWLREFQ KFAPALKIEP YYGSLQEREE 660
661 LRDILERNAG KYDVIVTTYN LAAGNKYDVS FLKNRNFNVV VYDEGHMLKN STSERFAKLM 720
721 KIRANFRLLL TGTPLQNNLK ELMSLLEFIM PNLFISKKES FDAIFKQRAK TTDDNKNHNP 780
781 LLAQEAITRA KTMMKPFILR RRKDQVLKHL PPKHTHIQYC ELNAIQKKIY DKEIQIVLEH 840
841 KRMIKDGELP KDAKEKSKLQ SSSSKNLIMA LRKASLHPLL FRNIYNDKII TKMSDAILDE 900
901 PAYAENGNKE YIKEDMSYMT DFELHKLCCN FPNTLSKYQL HNDEWMQSGK IDALKKLLKT 960
961 IIVDKQEKVL IFSLFTQVLD ILEMVLSTLD YKFLRLDGST QVNDRQLLID KFYEDKDIPI1020
1021 FILSTKAGGF GINLVCANNV IIFDQSFNPH DDRQAADRAH RVGQTKEVNI TTLITKDSIE1080
1081 EKIHQLAKNK LALDSYISED KKSQDVLESK VSDMLEDIIY DENSKPKGTK E |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
AAT2 FUN30 |
| View Details | Gavin AC, et al. (2002) |
|
FUN30 MIS1 TEL1 |
| View Details | Ho Y, et al. (2002) |
|
FUN30 PSK1 PSK2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
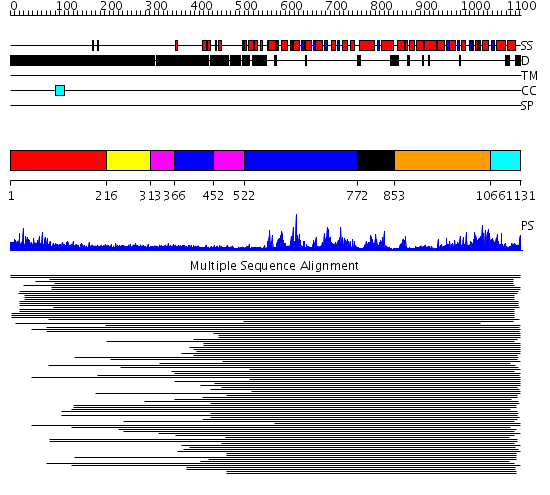
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 82.329997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [216..312] | 5.522879 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [313..365] [452..521] |
5.522879 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [366..451] [522..771] |
5.522879 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 5 | View Details | [772..852] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [853..1065] | 7.0 | Putative DEAD box RNA helicase | |
| 7 | View Details | [1066..1131] | 5.0 | Nucleotide excision repair enzyme UvrB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)