













| General Information: |
|
| Name(s) found: |
GZF3 /
YJL110C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IMP nitrogen compound metabolic process [TAS negative regulation of transcription [IDA regulation of nitrogen utilization [IGI |
| Molecular Function: |
transcription factor activity
[IDA specific RNA polymerase II transcription factor activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASQATTLRG YNIRKRDNVF EPKSSENLNS LNQSEEEGHI GRWPPLGYEA VSAEQKSAVQ 60
61 LRESQAGASI SNNMNFKAND KSFSTSTAGR MSPDTNSLHH ILPKNQVKNN GQTMDANCNN 120
121 NVSNDANVPV CKNCLTSTTP LWRRDEHGAM LCNACGLFLK LHGKPRPISL KTDVIKSRNR 180
181 KSNTNHAHNL DNFRNQTLIA ELKGDCNIES SGRKANRVTS EDKKKKSSQL LMGTSSTAKI 240
241 SKKPKTESKE RSDSHLSATK LEVLMSGDCS RPNLKPKLPK QDTAIYQEKL LTFPSYTDVK 300
301 EYSNSAHQSA FIKERSQFNA ASFPLNASHS VTSKTGADSP QLPHLSMLLG SLSSTSISNN 360
361 GSEIVSNCNN GIASTAATLA PTSSRTTDSN PSEVPNQIRS TMSSPDIISA KRNDPAPLSF 420
421 HMASINDMLE TRDRAISNVK TETTPPHFIP FLQSSKAPCI SKANSQSISN SVSSSDVSGR 480
481 KFENHPAKDL GDQLSTKLHK EEEIIKLKTR INELELVTDL YRRHINELDG KCRALEERLQ 540
541 RTVKQEGNKG G |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
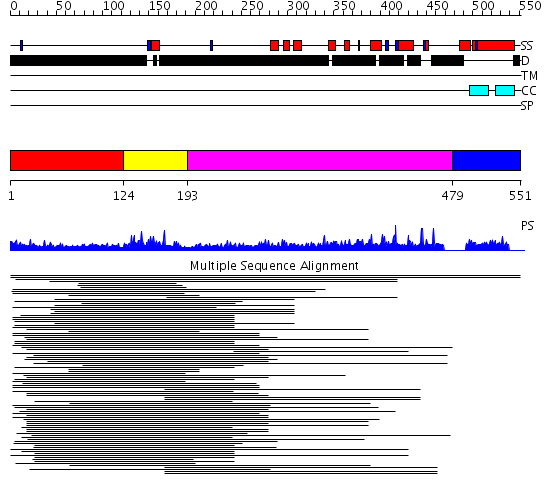
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [124..192] | 173.68867 | Erythroid transcription factor GATA-1 | |
| 3 | View Details | [193..478] | 19.197997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [479..551] | 1.003984 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)