













| General Information: |
|
| Name(s) found: |
FPR3 /
YML074C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 411 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
meiotic recombination checkpoint
[IDA |
| Molecular Function: |
peptidyl-prolyl cis-trans isomerase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDLLPLATY SLNVEPYTPV PAIDVTMPIT VRITMAALNP EAIDEENKPS TLRIIKRNPD 60
61 FEDDDFLGGD FDEDEIDEES SEEEEEEKTQ KKKKSKGKKA ESESEDDEED DDEDDEFQES 120
121 VLLTLSPEAQ YQQSLDLTIT PEEEVQFIVT GSYAISLSGN YVKHPFDTPM GVEGEDEDED 180
181 ADIYDSEDYD LTPDEDEIIG DDMDDLDDEE EEEVRIEEVQ EEDEEDNDGE EEQEEEEEEE 240
241 QKEEVKPEPK KSKKEKKRKH EEKEEEKKAK KVKKVEFKKD LEEGPTKPKS KKEQDKHKPK 300
301 SKVLEGGIVI EDRTIGDGPQ AKRGARVGMR YIGKLKNGKV FDKNTSGKPF AFKLGRGEVI 360
361 KGWDIGVAGM SVGGERRIII PAPYAYGKQA LPGIPANSEL TFDVKLVSMK N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
FPR3 GLC7 |
| View Details | Ho Y, et al. (2002) |

|
ADH2 ADH4 BMS1 CDC16 CDC25 CDC33 CDC53 CFT1 CIC1 CKA1 CKA2 CKB1 CKB2 CLU1 COF1 COR1 CTF4 DBP10 DBP2 DCP2 DED81 DIA2 DOG1 EDC3 EDE1 EGD1 ENP1 ENP2 ERB1 FET4 FIN1 FIP1 FPR3 FPR4 GLC7 GLC8 GPH1 GSY1 GSY2 HAS1 HER1 HHF1, HHF2 HHT1, HHT2 HOT1 HRR25 HTA1 HTB2 ILS1 KEL1 KRE33 KRI1 LOC1 LST4 MAM33 MCM2 MCM3 MCM5 MDH1 MDS3 MES1 MGM101 MHP1 MKT1 MMS4 MUS81 NHP2 NIP7 NMD3 NOG1 NOP12 NOP2 NOP4 NOP56 NOP7 NSA1 NUG1 PDC5 PDI1 PIM1 POB3 PRP8 PUF6 PWP1 QCR2 RAD53 REG1 RPC10 RPF2 RPM2 RRP12 RRP17 SAP185 SAP190 SCD5 SDS22 SEC23 SEH1 SEN1 SFB3 SGM1 SIT4 SKP1 SLT2 SNP1 SPB1 SPT16 SRP1 SSF1 SSF2 SSZ1 SUI2 SUI3 TAF14 TGL1 TIF6 TSR1 UBI4 URB1 UTP10 UTP22 VAC14 YAK1 YBL104C YER084W YHB1 YPI1 YRA1 YTM1 |
| View Details | Qiu et al. (2008) |

|
FPR3 GAR1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
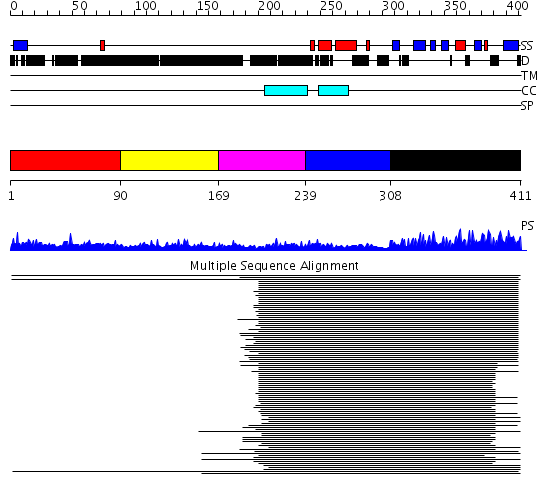
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 8.75 | No description for 1l8mA was found. | |
| 2 | View Details | [90..168] | 8.75 | No description for 1l8mA was found. | |
| 3 | View Details | [169..238] | 8.75 | No description for 1l8mA was found. | |
| 4 | View Details | [239..307] | 8.75 | No description for 1l8mA was found. | |
| 5 | View Details | [308..411] | 250.218487 | Calcineurin (FKBP12.6) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)