













| General Information: |
|
| Name(s) found: |
MGM101 /
YJR144W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 269 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial chromosome
[IDA mitochondrion [IDA mitochondrial nucleoid [IDA |
| Biological Process: |
DNA repair
[IMP mitochondrial genome maintenance [IMP |
| Molecular Function: |
DNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSIFKVRGC VSHAAQFCQK RTVVSTGTSN TATAGAVRKS FNSTETKPVF ATKSEAGNGS 60
61 HMKEYSSGIN SKLGGTPLET RSTADDSLNN SYKQVKGDID WYTSWYGLGM KPFEAKVQKD 120
121 LIEPLDPKDI EIKPDGLIYL PEIKYRRILN KAFGAGGWGL VPRSQTIVTS KLVTREYGLI 180
181 CHGQLISVAR GEQDYFNEAG IPTATEGCKS NALMRCCKDL GVGSELWDPV FIKKFKVDHC 240
241 TEKFVEHVTT KRKKKIWLRK DRQVEYPYK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
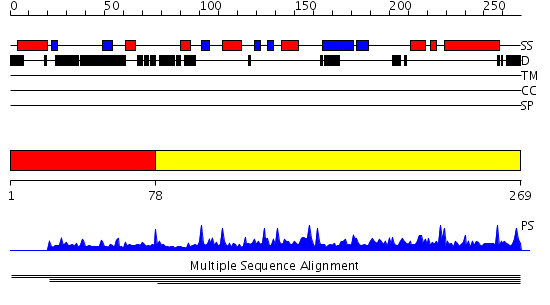
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [78..269] | 1.003908 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)