













| General Information: |
|
| Name(s) found: |
MPH1 /
YIR002C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 993 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
DNA repair
[IMP |
| Molecular Function: |
RNA helicase activity
[IMP 3'-5' DNA helicase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASADDYFSD FEDDELDKLY EKAINKSVKE TITRRAVPVQ KDLHDNVLPG QKTVYEEIQR 60
61 DVSFGPTHHE LDYDALSFYV YPTNYEVRDY QYTIVHKSLF QNTLCAIPTG MGKTFIASTV 120
121 MLNYFRWTKK AKIIFTAPTR PLVAQQIKAC LGITGIPSDQ TAILLDKSRK NREEIWANKR 180
181 VFFATPQVVE NDLKRGVLDP KDIVCLVIDE AHRATGSSAY TNVVKFIDRF NSSYRLLALT 240
241 ATPASDLEGV QEVVNNLDIS KIEIRTEESM DIVKYMKKRK KEKIEVPLLL EIEDIIEQLG 300
301 MAVKPVLQQA IELGIYEECD PSQINAFKAM QQSQKIIANP TIPEGIKWRN FFILQLLNNV 360
361 GQMLKRLKIY GIRTFFNYFQ NKCTEFTTKY NLKKSTNKIA AEFYYHPILK NIKNQCENYL 420
421 SDPKFVGHGK LQCVRDELMD FFQKRGSDSR VIIFTELRES ALEIVKFIDS VADDQIRPHI 480
481 FIGQARAKEG FDEVKYTRKH APKGRKKVER LHRQEQEKFL EAERTKRAAN DKLERSARRT 540
541 GSSEEAQISG MNQKMQKEVI HNFKKGEYNV LVCTSIGEEG LDIGEVDLII CYDTTSSPIK 600
601 NIQRMGRTGR KRDGKIVLLF SSNESYKFER AMEDYSTLQA LISKQCIDYK KSDRIIPEDI 660
661 IPECHETLIT INDENEIINE MEDVDEVIRY ATQCMMGKKV KPKKAITKKK RVQENKKPKK 720
721 FFMPDNVETS IVSASTLINK FLVNESGGKQ LVTSNENPSK KRKIFKALDN LENDSTEEAS 780
781 SSLETEDEEV SDDNNVFIAE GQNGCQKDLE TAIIRTGESL TTLKPLHNFE RPNMALFVND 840
841 CGLPTKIEKN VKDIRGNQHN LEKEKSCTVD KNNMVLSLDD WNFFRNRYIP EGVSFDVEPN 900
901 FVQYTKGVKV PHCHKVSKII TLFNDESNDN KKRTIDMNYT KCLARGMLRD EKKFVKVNDK 960
961 SQVDNNSVNH DSSQSFTLSN AELDDILGSD SDF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
MGM101 MPH1 MSH2 MSH6 RAD52 RFA1 RFA2 |
| View Details | Gavin AC, et al. (2006) |
|
MPH1 MSH6 RAD52 RFA1 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 ECM29 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MRPS28 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 RSC8 RVB2 SGS1 SHS1 SRO7 SSL1 TFA1 TFB1 TFB4 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |
|
AI1 CDC28 CEG1 CKA1 CKA2 CKB1 CKB2 FKH1 FUN12 FYV8 GCD2 GCD7 GCN3 HHF1, HHF2 HHO1 HTB1 IMD2 IMD3 IMD4 MAG1 MBP1 MGM101 MPH1 MSH2 NET1 NOP1 NOP12 RET1 RPC82 RRP1 SEC2 SIN3 SUI2 SUI3 TIF4631 TIF4632 UBP12 URE2 UTP22 YGR017W YMR144W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
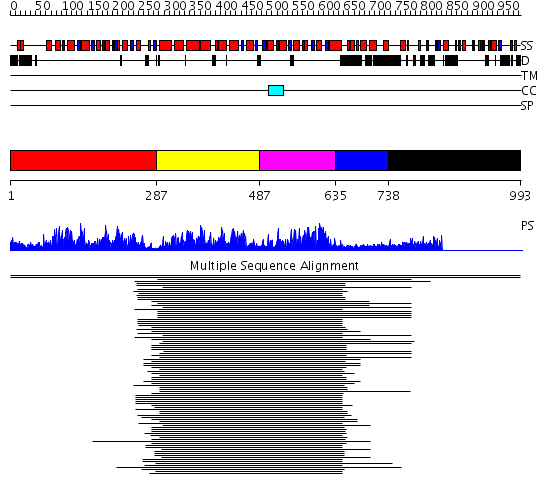
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | 68.69897 | Initiation factor 4a | |
| 2 | View Details | [287..486] | 145.228787 | Putative DEAD box RNA helicase | |
| 3 | View Details | [487..634] | 145.228787 | Putative DEAD box RNA helicase | |
| 4 | View Details | [635..737] | 65.522879 | Nucleotide excision repair enzyme UvrB | |
| 5 | View Details | [738..993] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)