













| General Information: |
|
| Name(s) found: |
MLH1 /
YMR167W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 769 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IMP mitochondrion [IDA |
| Biological Process: |
reciprocal meiotic recombination
[TAS meiotic mismatch repair [IMP |
| Molecular Function: |
ATPase activity
[ISS DNA binding [IDA ATP binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLRIKALDA SVVNKIAAGE IIISPVNALK EMMENSIDAN ATMIDILVKE GGIKVLQITD 60
61 NGSGINKADL PILCERFTTS KLQKFEDLSQ IQTYGFRGEA LASISHVARV TVTTKVKEDR 120
121 CAWRVSYAEG KMLESPKPVA GKDGTTILVE DLFFNIPSRL RALRSHNDEY SKILDVVGRY 180
181 AIHSKDIGFS CKKFGDSNYS LSVKPSYTVQ DRIRTVFNKS VASNLITFHI SKVEDLNLES 240
241 VDGKVCNLNF ISKKSISPIF FINNRLVTCD LLRRALNSVY SNYLPKGNRP FIYLGIVIDP 300
301 AAVDVNVHPT KREVRFLSQD EIIEKIANQL HAELSAIDTS RTFKASSIST NKPESLIPFN 360
361 DTIESDRNRK SLRQAQVVEN SYTTANSQLR KAKRQENKLV RIDASQAKIT SFLSSSQQFN 420
421 FEGSSTKRQL SEPKVTNVSH SQEAEKLTLN ESEQPRDANT INDNDLKDQP KKKQKLGDYK 480
481 VPSIADDEKN ALPISKDGYI RVPKERVNVN LTSIKKLREK VDDSIHRELT DIFANLNYVG 540
541 VVDEERRLAA IQHDLKLFLI DYGSVCYELF YQIGLTDFAN FGKINLQSTN VSDDIVLYNL 600
601 LSEFDELNDD ASKEKIISKI WDMSSMLNEY YSIELVNDGL DNDLKSVKLK SLPLLLKGYI 660
661 PSLVKLPFFI YRLGKEVDWE DEQECLDGIL REIALLYIPD MVPKVDTSDA SLSEDEKAQF 720
721 INRKEHISSL LEHVLFPCIK RRFLAPRHIL KDVVEIANLP DLYKVFERC |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
MLH1 PMS1 |
| View Details | Krogan NJ, et al. (2006) |

|
MLH1 MLH2 |
| View Details | Gavin AC, et al. (2006) |

|
MLH1 PMS1 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |
|
ISN1 MGE1 MLH1 RAD51 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
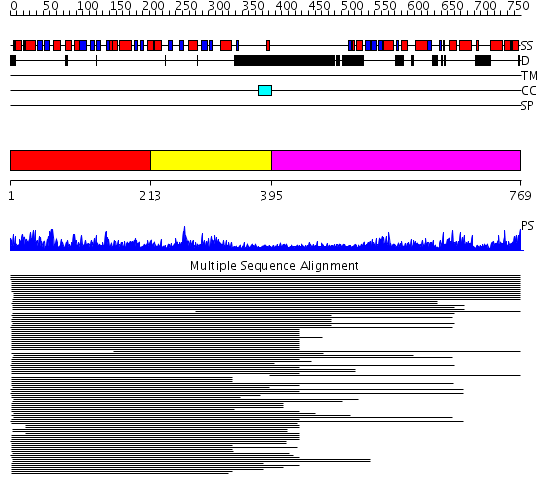
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..212] | 550.0 | DNA mismatch repair protein MutL | |
| 2 | View Details | [213..394] | 550.0 | DNA mismatch repair protein MutL | |
| 3 | View Details | [395..769] | 1.300902 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)