













| General Information: |
|
| Name(s) found: |
SIN3 /
YOL004W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1536 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Sin3 complex
[IPI histone deacetylase complex [IPI |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IMP chromatin silencing at rDNA [IMP telomere maintenance [IMP double-strand break repair via nonhomologous end joining [IMP histone deacetylation [IMP chromatin silencing at silent mating-type cassette [IMP positive regulation of transcription from RNA polymerase II promoter [IMP chromatin silencing at telomere [IMP |
| Molecular Function: |
transcription coactivator activity
[IMP histone deacetylase activity [IMP transcription corepressor activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQVWHNSNS QSNDVATSND ATGSNERNEK EPSLQGNKPG FVQQQQRITL PSLSALSTKE 60
61 EDRRDSNGQQ ALTSHAAHIL GYPPPHSNAM PSIATDSALK QPHEYHPRPK SSSSSPSINA 120
121 SLMNAGPAPL PTVGAASFSL SRFDNPLPIK APVHTEEPKS YNGLQEEEKA TQRPQDCKEV 180
181 PAGVQPADAP DPSSNHADAN DDNNNNENSH DEDADYRPLN VKDALSYLEQ VKFQFSSRPD 240
241 IYNLFLDIMK DFKSQAIDTP GVIERVSTLF RGYPILIQGF NTFLPQGYRI ECSSNPDDPI 300
301 RVTTPMGTTT VNNNISPSGR GTTDAQELGS FPESDGNGVQ QPSNVPMVPS SVYQSEQNQD 360
361 QQQSLPLLAT SSGLPSIQQP EMPAHRQIPQ SQSLVPQEDA KKNVDVEFSQ AISYVNKIKT 420
421 RFADQPDIYK HFLEILQTYQ REQKPINEVY AQVTHLFQNA PDLLEDFKKF LPDSSASANQ 480
481 QVQHAQQHAQ QQHEAQMHAQ AQAQAQAQAQ VEQQKQQQQF LYPASGYYGH PSNRGIPQQN 540
541 LPPIGSFSPP TNGSTVHEAY QDQQHMQPPH FMPLPSIVQH GPNMVHQGIA NENPPLSDLR 600
601 TSLTEQYAPS SIQHQQQHPQ SISPIANTQY GDIPVRPEID LDPSIVPVVP EPTEPIENNI 660
661 SLNEEVTFFE KAKRYIGNKH LYTEFLKILN LYSQDILDLD DLVEKVDFYL GSNKELFTWF 720
721 KNFVGYQEKT KCIENIVHEK HRLDLDLCEA FGPSYKRLPK SDTFMPCSGR DDMCWEVLND 780
781 EWVGHPVWAS EDSGFIAHRK NQYEETLFKI EEERHEYDFY IESNLRTIQC LETIVNKIEN 840
841 MTENEKANFK LPPGLGHTSM TIYKKVIRKV YDKERGFEII DALHEHPAVT APVVLKRLKQ 900
901 KDEEWRRAQR EWNKVWRELE QKVFFKSLDH LGLTFKQADK KLLTTKQLIS EISSIKVDQT 960
961 NKKIHWLTPK PKSQLDFDFP DKNIFYDILC LADTFITHTT AYSNPDKERL KDLLKYFISL1020
1021 FFSISFEKIE ESLYSHKQNV SESSGSDDGS SIASRKRPYQ QEMSLLDILH RSRYQKLKRS1080
1081 NDEDGKVPQL SEPPEEEPNT IEEEELIDEE AKNPWLTGNL VEEANSQGII QNRSIFNLFA1140
1141 NTNIYIFFRH WTTIYERLLE IKQMNERVTK EINTRSTVTF AKDLDLLSSQ LSEMGLDFVG1200
1201 EDAYKQVLRL SRRLINGDLE HQWFEESLRQ AYNNKAFKLY TIDKVTQSLV KHAHTLMTDA1260
1261 KTAEIMALFV KDRNASTTSA KDQIIYRLQV RSHMSNTENM FRIEFDKRTL HVSIQYIALD1320
1321 DLTLKEPKAD EDKWKYYVTS YALPHPTEGI PHEKLKIPFL ERLIEFGQDI DGTEVDEEFS1380
1381 PEGISVSTLK IKIQPITYQL HIENGSYDVF TRKATNKYPT IANDNTQKGM VSQKKELISK1440
1441 FLDCAVGLRN NLDEAQKLSM QKKWENLKDS IAKTSAGNQG IESETEKGKI TKQEQSDNLD1500
1501 SSTASVLPAS ITTVPQDDNI ETTGNTESSD KGAKIQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
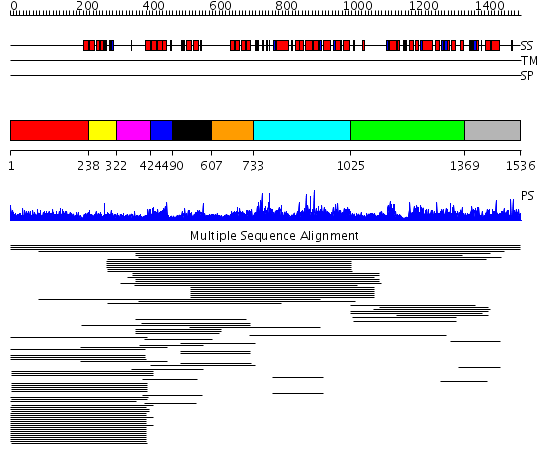
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | 11.5 | RBP1 | |
| 2 | View Details | [238..321] | 19.346787 | Paired amphipathic helix repeat No confident structure predictions are available. | |
| 3 | View Details | [322..423] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [424..489] | 19.69897 | Paired amphipathic helix repeat No confident structure predictions are available. | |
| 5 | View Details | [490..606] | 172.134302 | Sin3A | |
| 6 | View Details | [607..732] | 11.33 | Sin3A | |
| 7 | View Details | [733..1024] | 1.021966 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1025..1368] | 2.019929 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1369..1536] | 2.014993 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1179..1355] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1356..1449] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1450..1536] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)