













| General Information: |
|
| Name(s) found: |
STB1 /
YNL309W
[SGD]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 420 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA Sin3 complex [IPI |
| Biological Process: |
G1/S transition of mitotic cell cycle
[IEP |
| Molecular Function: |
transcription activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQPQMSPEK EQELASKILH RAELAQMTRQ LKLGLSNVPS TKRKQDSTTK KRSGEDAEDV 60
61 DEDHKTLLEA ISPAKKPLHD DTNKMTVISP VKFVEKPNTP PSSRQRKAED RSQQIKPRKE 120
121 DTPSTPRASA TPIILPHASS HYQRPHDKNF MTPKRNNNNS SNHSNNNNNI KKKAAGSKDA 180
181 PQDSDNTAGA DLLMYLATSP YNKSSHHGTP MAVRMPTTPR SYHYASQLSL NGNTASTSND 240
241 AVRFSHIKPS ASSPQSTFKS NLLPNFPDES LMDSPSLYLS NNNGSVQATL SPQQRRKPTT 300
301 NTLHPPSNVP TTPSRELNGT NFNLLRTPNF NMGDYLHNLF SPSPRVPAQQ GASNTSASIP 360
361 SVPAMVPGSS SNTSAIATAA ISSHTTNNFL DMNANGIPLI VGPGTDRIGE GESIDDKLTD 420
421 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
RPD3 SIN3 STB1 STB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
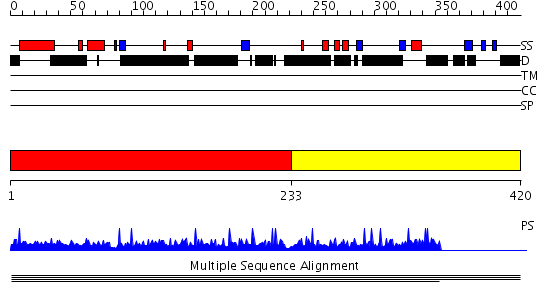
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..232] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [233..420] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)