













| General Information: |
|
| Name(s) found: |
EAF3 /
YPR023C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 401 amino acids |
Gene Ontology: |
|
| Cellular Component: |
NuA4 histone acetyltransferase complex
[IPI histone acetyltransferase complex [IDA |
| Biological Process: |
DNA repair
[IDA histone acetylation [IDA histone deacetylation [IDA regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
histone acetyltransferase activity
[IDA histone deacetylase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK 60
61 LGEDESIPEE IINGKCFFIH YQGWKSSWDE WVGYDRIRAY NEENIAMKKR LANEAKEAKK 120
121 SLLEQQKKKK LSTSLGGPSN GGKRKGDSRS NASISKSTSQ SFLTSSVSGR KSGRSSANSL 180
181 HPGSSLRSSS DQNGNDDRRR SSSLSPNMLH HIAGYPTPKI SLQIPIKLKS VLVDDWEYVT 240
241 KDKKICRLPA DVTVEMVLNK YEHEVSQELE SPGSQSQLSE YCAGLKLYFD KCLGNMLLYR 300
301 LERLQYDELL KKSSKDQKPL VPIRIYGAIH LLRLISVLPE LISSTTMDLQ SCQLLIKQTE 360
361 DFLVWLLMHV DEYFNDKDPN RSDDALYVNT SSQYEGVALG M |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
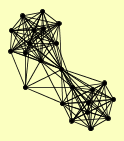
|
ARP4 CTI6 DEP1 EAF1 EAF3 EAF5 EAF6 EAF7 EPL1 ESA1 PHO23 RCO1 RPD3 RXT2 SAP30 SDS3 SIN3 SRP1 SWC4 TRA1 UME1 YAF9 YNG2 |
| View Details | Krogan NJ, et al. (2006) |
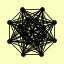
|
ASH1 COQ6 CTI6 DEP1 EAF3 GRX5 PHO23 RCO1 RPD3 RXT2 RXT3 SAP30 SAY1 SDS3 SIN3 UME1 |
| View Details | Gavin AC, et al. (2002) |
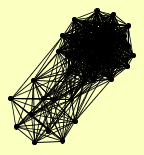
|
ABF2 ACT1 ADH1 ARP4 CKA1 CKA2 CKB1 CKB2 CTI6 DEP1 DOT6 EAF1 EAF3 EAF5 ECM16 EPL1 ESA1 HHF1, HHF2 HTA2 IOC3 ISW1 MED4 NHP10 NRD1 PHO23 RCO1 RPD3 RSC8 RXT2 RXT3 SAP30 SDS3 SIN3 STO1 TOP1 TRA1 TUP1 UME1 UME6 VPS1 YAF9 YAP1 |
| View Details | Ho Y, et al. (2002) |

|
DIS3 EAF3 FIP1 HAS1 HPR1 KAP95 MES1 MFT1 NAM8 NOP6 NUP1 NUP2 NUP60 PAP1 PCT1 REB1 RNT1 RRP4 RRP43 RRP6 RTT103 SIF2 SIN3 SNU56 SRP1 STO1 THO2 TRA1 UME1 YPR090W |
| View Details | Qiu et al. (2008) |
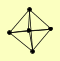
|
ARP4 EAF3 EPL1 SGF73 TRA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
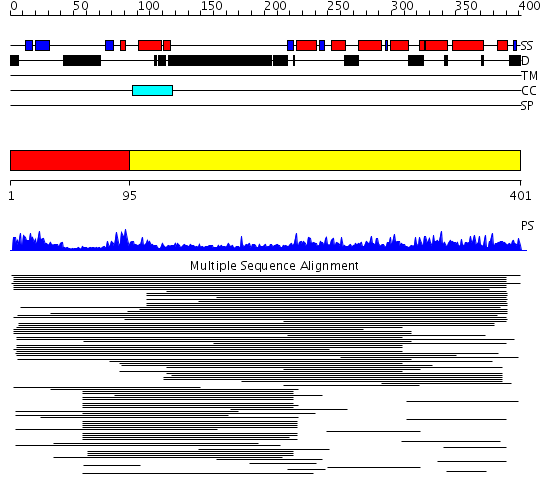
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [95..401] | 10.057965 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)