













| General Information: |
|
| Name(s) found: |
THO2 /
YNL139C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1597 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC THO complex [IMP |
| Biological Process: |
mRNA export from nucleus
[IMP DNA recombination [IMP RNA elongation from RNA polymerase II promoter [IMP |
| Molecular Function: |
nucleic acid binding
[IDA DNA binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEQTLLSKL NALSQKVIPP ASPSQASILT EEVIRNWPER SKTLCSDFTA LESNDEKEDW 60
61 LRTLFIELFD FINKNDENSP LKLSDVASFT NELVNHERQV SQASIVGKMF IAVSSTVPNI 120
121 NDLTTISLCK LIPSLHEELF KFSWISSKLL NKEQTTLLRH LLKKSKYELK KYNLLVENSV 180
181 GYGQLVALLI LAYYDPDNFS KVSAYLKEIY HIMGKYSLDS IRTLDVILNV SSQFITEGYK 240
241 FFIALLRKSD SWPSSHVANN SNYSSLNEGG NMIAANIISF NLSQYNEEVD KENYERYMDM 300
301 CCILLKNGFV NFYSIWDNVK PEMEFLQEYI QNLETELEEE STKGVENPLA MAAALSTENE 360
361 TDEDNALVVN DDVNMKDKIS EETNADIESK GKQKTQQDIL LFGKIKLLER LLIHGCVIPV 420
421 IHVLKQYPKV LYVSESLSRY LGRVFEYLLN PLYTSMTSSG ESKDMATALM ITRIDNGILA 480
481 HKPRLIHKYK THEPFESLEL NSSYVFYYSE WNSNLTPFAS VNDLFENSHI YLSIIGPYLG 540
541 RIPTLLSKIS RIGVADIQKN HGSESLHVTI DKWIDYVRKF IFPATSLLQN NPIATSEVYE 600
601 LMKFFPFEKR YFIYNEMMTK LSQDILPLKV SFNKAEREAK SILKALSIDT IAKESRRFAK 660
661 LISTNPLASL VPAVKQIENY DKVSELVVYT TKYFNDFAYD VLQFVLLLRL TYNRPAVQFD 720
721 GVNQAMWVQR LSIFIAGLAK NCPNMDISNI ITYILKTLHN GNIIAVSILK ELIITVGGIR 780
781 DLNEVNMKQL LMLNSGSPLK QYARHLIYDF RDDNSVISSR LTSFFTDQSA ISEIILLLYT 840
841 LNLKANTQNS HYKILSTRCD EMNTLLWSFI ELIKHCLKGK AFEENVLPFV ELNNRFHLST 900
901 PWTFHIWRDY LDNQLNSNEN FSIDELIEGA EFSDVDLTKI SKDLFTTFWR LSLYDIHFDK 960
961 SLYDERKNAL SGENTGHMSN RKKHLIQNQI KDILVTGISH QRAFKKTSEF ISEKSNVWNK1020
1021 DCGEDQIKIF LQNCVVPRVL FSPSDALFSS FFIFMAFRTE NLMSILNTCI TSNILKTLLF1080
1081 CCTSSEAGNL GLFFTDVLKK LEKMRLNGDF NDQASRKLYE WHSVITEQVI DLLSEKNYMS1140
1141 IRNGIEFMKH VTSVFPVVKA HIQLVYTTLE ENLINEERED IKLPSSALIG HLKARLKDAL1200
1201 ELDEFCTLTE EEAEQKRIRE MELEEIKNYE TACQNEQKQV ALRKQLELNK SQRLQNDPPK1260
1261 SVASGSAGLN SKDRYTYSRN EPVIPTKPSS SQWSYSKVTR HVDDINHYLA TNHLQKAISL1320
1321 VENDDETRNL RKLSKQNMPI FDFRNSTLEI FERYFRTLIQ NPQNPDFAEK IDSLKRYIKN1380
1381 ISREPYPDTT SSYSEAAAPE YTKRSSRYSG NAGGKDGYGS SNYRGPSNDR SAPKNIKPIS1440
1441 SYAHKRSELP TRPSKSKTYN DRSRALRPTG PDRGDGFDQR DNRLREEYKK NSSQRSQLRF1500
1501 PEKPFQEGKD SSKANPYQAS SYKRDSPSEN EEKPNKRFKK DETIRNKFQT QDYRNTRDSG1560
1561 AAHRANENQR YNGNRKSNTQ ALPQGPKGGN YVSRYQR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
GBP2 HPR1 HRB1 MFT1 SUB2 TEX1 THO2 THP2 |
| View Details | Krogan NJ, et al. (2006) |
|
CPR7 GBP2 GVP36 HHO1 HPR1 HRB1 MFT1 SUB2 TEX1 THO2 THP2 ULS1 YNL140C |
| View Details | Gavin AC, et al. (2006) |
|
HPR1 HRB1 MFT1 SUB2 TEX1 THO2 THP2 |
| View Details | Gavin AC, et al. (2002) |

|
AIR2 BRR1 BUR2 CBC2 HTA1 KAP95 LUC7 MSH4 MUD1 NAB3 NAB6 NAM8 NPL3 NRD1 NUP60 PRP39 PRP40 PRP42 RSE1 SCP160 SEN1 SGV1 SMD1 SMD2 SMD3 SNP1 SNU56 SNU71 SRO9 SRP1 STO1 THO2 TIF4631 TIF4632 YRA2 |
| View Details | Ho Y, et al. (2002) |
|
AHA1 BCY1 CAR2 CYS4 DIS3 EAF3 FIP1 GBP2 GND1 GTO1 HAS1 HPR1 IMD3 KAP95 LAA1 LYS1 MES1 MFT1 NAM8 NOP6 NUP1 NUP2 NUP60 PAP1 PCT1 PRB1 REB1 RNT1 RRP4 RRP43 RRP6 RTT103 SAH1 SIF2 SIN3 SNO2 SNU56 SRP1 SSK2 SSN8 STO1 SUB2 TEX1 THI22 THO2 THP2 TIF1, TIF2 TPK2 TPK3 TRA1 UME1 VPS21 YBR028C YCK1 YCK2 YCK3 YGR111W YPR090W YPT53 YRA1 |
| View Details | Qiu et al. (2008) |
|
HPR1 MFT1 SUB2 THO2 THP2 YRA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
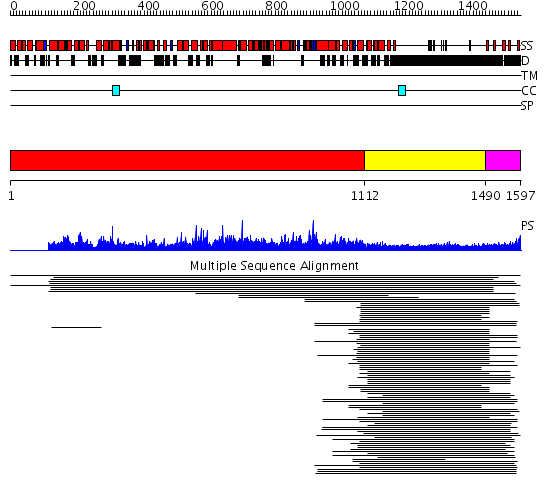
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1111] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [1112..1489] | 22.39794 | Heavy meromyosin subfragment | |
| 3 | View Details | [1490..1597] | 2.662996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [515..743] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [744..958] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [959..1087] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1088..1514] | 23.0 | Heavy meromyosin subfragment | |
| 8 | View Details | [1515..1597] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)