













| General Information: |
|
| Name(s) found: |
NUP1 /
YOR098C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1076 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
protein export from nucleus
[TAS ribosomal protein import into nucleus [TAS mRNA export from nucleus [IMP rRNA export from nucleus [TAS nuclear envelope organization [IMP RNA export from nucleus [IMP nuclear pore organization [TAS snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS protein import into nucleus [IMP tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS |
| Molecular Function: |
protein binding
[IPI structural molecule activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSNTSSVMS SPRVEKRSFS STLKSFFTNP NKKRPSSKKV FSSNLSYANH LEESDVEDTL 60
61 HVNKRKRVSG TSQHSDSLTQ NNNNAPIIIY GTENTERPPL LPILPIQRLR LLREKQRVRN 120
121 MRELGLIQST EFPSITSSVI LGSQSKSDEG GSYLCTSSTP SPIKNGSCTR QLAGKSGEDT 180
181 NVGLPILKSL KNRSNRKRFH SQSKGTVWSA NFEYDLSEYD AIQKKDNKDK EGNAGGDQKT 240
241 SENRNNIKSS ISNGNLATGP NLTSEIEDLR ADINSNRLSN PQKNLLLKGP ASTVAKTAPI 300
301 QESFVPNSER SGTPTLKKNI EPKKDKESIV LPTVGFDFIK DNETPSKKTS PKATSSAGAV 360
361 FKSSVEMGKT DKSTKTAEAP TLSFNFSQKA NKTKAVDNTV PSTTLFNFGG KSDTVTSASQ 420
421 PFKFGKTSEK SENHTESDAP PKSTAPIFSF GKQEENGDEG DDENEPKRKR RLPVSEDTNT 480
481 KPLFDFGKTG DQKETKKGES EKDASGKPSF VFGASDKQAE GTPLFTFGKK ADVTSNIDSS 540
541 AQFTFGKAAT AKETHTKPSE TPATIVKKPT FTFGQSTSEN KISEGSAKPT FSFSKSEEER 600
601 KSSPISNEAA KPSFSFPGKP VDVQAPTDDK TLKPTFSFTE PAQKDSSVVS EPKKPSFTFA 660
661 SSKTSQPKPL FSFGKSDAAK EPPGSNTSFS FTKPPANETD KRPTPPSFTF GGSTTNNTTT 720
721 TSTKPSFSFG APESMKSTAS TAAANTEKLS NGFSFTKFNH NKEKSNSPTS FFDGSASSTP 780
781 IPVLGKPTDA TGNTTSKSAF SFGTANTNGT NASANSTSFS FNAPATGNGT TTTSNTSGTN 840
841 IAGTFNVGKP DQSIASGNTN GAGSAFGFSS SGTAATGAAS NQSSFNFGNN GAGGLNPFTS 900
901 ATSSTNANAG LFNKPPSTNA QNVNVPSAFN FTGNNSTPGG GSVFNMNGNT NANTVFAGSN 960
961 NQPHQSQTPS FNTNSSFTPS TVPNINFSGL NGGITNTATN ALRPSDIFGA NAASGSNSNV1020
1021 TNPSSIFGGA GGVPTTSFGQ PQSAPNQMGM GTNNGMSMGG GVMANRKIAR MRHSKR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
KAP95 NUP1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GLE2 NOP14 NUP1 NUP100 NUP116 NUP133 NUP157 NUP159 NUP170 NUP188 NUP2 NUP42 POM152 |
| View Details | Krogan NJ, et al. (2006) |
|
CBC2 KAP95 NUP1 NUP2 NUP60 OGG1 PCT1 RAD53 SRP1 STO1 ULP1 URB2 YRA2 |
| View Details | Ho Y, et al. (2002) |

|
DIS3 EAF3 FIP1 HAS1 HPR1 KAP95 MES1 MFT1 NAM8 NOP6 NUP1 NUP2 NUP60 PAP1 PCT1 REB1 RNT1 RRP4 RRP43 RRP6 RTT103 SIF2 SIN3 SNU56 SRP1 STO1 THO2 TRA1 UME1 YPR090W |
| View Details | Qiu et al. (2008) |
|
ASM4 DBP5 GLE2 KAP122 KAP123 MEX67 MTR2 NDC1 NIC96 NSP1 NUP1 NUP100 NUP116 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP192 NUP2 NUP42 NUP49 NUP53 NUP57 NUP60 NUP82 NUP84 NUP85 POM152 SEC13 SUS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
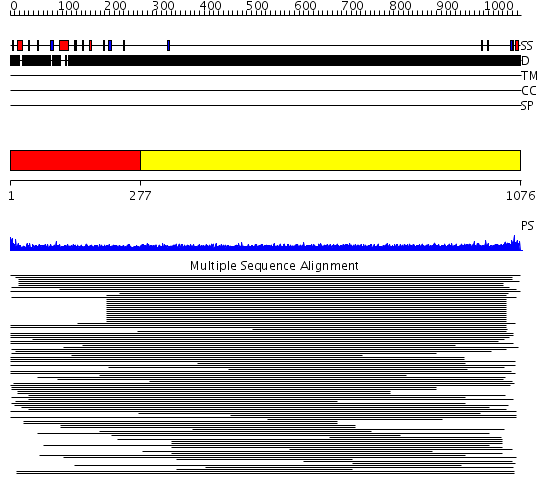
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..276] | 1.150997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [277..1076] | 10.232997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [389..632] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [633..746] | 2.214988 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [747..892] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [893..1076] | 2.19299 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)