













| General Information: |
|
| Name(s) found: |
MRS6 /
YOR370C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 603 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA membrane [IDA |
| Biological Process: |
intracellular protein transport
[TAS |
| Molecular Function: |
GTPase activator activity
[IEA]
Rab GTPase binding [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSPERRPSM AERRPSFFSF TQNPSPLVVP HLAGIEDPLP ATTPDKVDVL IAGTGMVESV 60
61 LAAALAWQGS NVLHIDKNDY YGDTSATLTV DQIKRWVNEV NEGSVSCYKN AKLYVSTLIG 120
121 SGKYSSRDFG IDLSPKILFA KSDLLSILIK SRVHQYLEFQ SLSNFHTYEN DCFEKLTNTK 180
181 QEIFTDQNLP LMTKRNLMKF IKFVLNWEAQ TEIWQPYAER TMSDFLGEKF KLEKPQVFEL 240
241 IFSIGLCYDL NVKVPEALQR IRRYLTSFDV YGPFPALCSK YGGPGELSQG FCRSAAVGGA 300
301 TYKLNEKLVS FNPTTKVATF QDGSKVEVSE KVIISPTQAP KDSKHVPQQQ YQVHRLTCIV 360
361 ENPCTEWFNE GESAAMVVFP PGSLKSGNKE VVQAFILGAG SEICPEGTIV WYLSTTEQGP 420
421 RAEMDIDAAL EAMEMALLRE SSSGLENDEE IVQLTGNGHT IVNSVKLGQS FKEYVPRERL 480
481 QFLFKLYYTQ YTSTPPFGVV NSSFFDVNQD LEKKYIPGAS DNGVIYTTMP SAEISYDEVV 540
541 TAAKVLYEKI VGSDDDFFDL DFEDEDEIQA SGVANAEQFE NAIDDDDDVN MEGSGEFVGE 600
601 MEI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
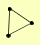
|
GDI1 MRS6 VPS21 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
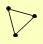
|
BET2 BET4 MRS6 |
| View Details | Krogan NJ, et al. (2006) |
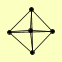
|
GDI1 IDP3 MRS6 VPS21 YPT6 |
| View Details | Gavin AC, et al. (2006) |

|
MRS6 PYK2 |
| View Details | Ho Y, et al. (2002) |
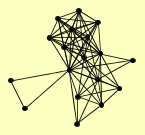
|
BCY1 CDC33 DSS4 GDI1 JJJ1 LAS1 MRS6 POR1 SEC4 SFP1 TPK1 TPK3 TPS1 VPS21 YPT1 YPT10 YPT52 YPT53 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
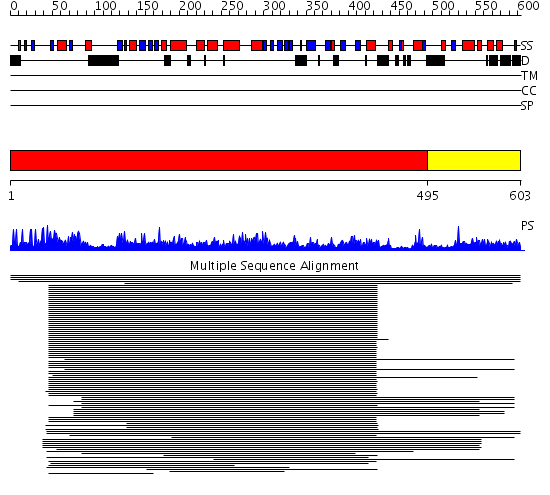
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..494] | 418.228787 | Guanine nucleotide dissosiation inhibitor, GDI | |
| 2 | View Details | [495..603] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)