













| General Information: |
|
| Name(s) found: |
JJJ1 /
YNL227C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 590 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA cytosol [IDA |
| Biological Process: |
ribosomal large subunit biogenesis
[IMP regulation of cell size [IMP endocytosis [IMP |
| Molecular Function: |
Hsp70/Hsc70 protein regulator activity
[IDA ATPase activator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTCYYELLG VETHASDLEL KKAYRKKALQ YHPDKNPDNV EEATQKFAVI RAAYEVLSDP 60
61 QERAWYDSHK EQILNDTPPS TDDYYDYEVD ATVTGVTTDE LLLFFNSALY TKIDNSAAGI 120
121 YQIAGKIFAK LAKDEILSGK RLGKFSEYQD DVFEQDINSI GYLKACDNFI NKTDKLLYPL 180
181 FGYSPTDYEY LKHFYKTWSA FNTLKSFSWK DEYMYSKNYD RRTKREVNRR NEKARQQARN 240
241 EYNKTVKRFV VFIKKLDKRM KEGAKIAEEQ RKLKEQQRKN ELNNRRKFGN DNNDEEKFHL 300
301 QSWQTVKEEN WDELEKVYDN FGEFENSKND KEGEVLIYEC FICNKTFKSE KQLKNHINTK 360
361 LHKKNMEEIR KEMEEENITL GLDNLSDLEK FDSADESVKE KEDIDLQALQ AELAEIERKL 420
421 AESSSEDESE DDNLNIEMDI EVEDVSSDEN VHVNTKNKKK RKKKKKAKVD TETEESESFD 480
481 DTKDKRSNEL DDLLASLGDK GLQTDDDEDW STKAKKKKGK QPKKNSKSTK STPSLSTLPS 540
541 SMSPTSAIEV CTTCGESFDS RNKLFNHVKI AGHAAVKNVV KRKKVKTKRI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
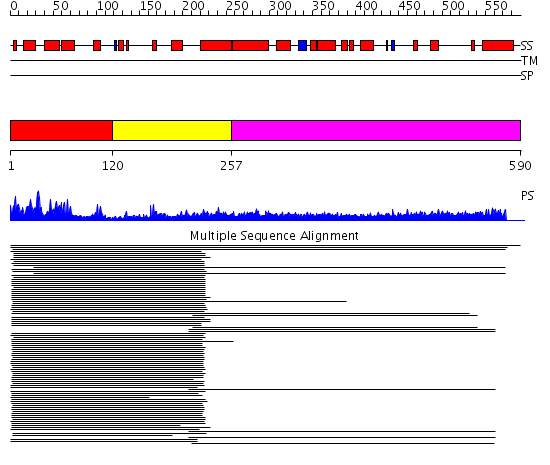
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | 119.512579 | DnaJ chaperone, N-terminal (J) domain | |
| 2 | View Details | [120..256] | 9.69897 | Cysteine-rich domain of the chaperone protein DnaJ. | |
| 3 | View Details | [257..590] | 10.0 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)