













| General Information: |
|
| Name(s) found: |
PYK2 /
YOR347C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 506 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA cytosol [TAS] |
| Biological Process: |
pyruvate metabolic process
[IMP |
| Molecular Function: |
pyruvate kinase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPESRLQRLA NLKIGTPQQL RRTSIIGTIG PKTNSCEAIT ALRKAGLNII RLNFSHGSYE 60
61 FHQSVIENAV KSEQQFPGRP LAIALDTKGP EIRTGRTLND QDLYIPVDHQ MIFTTDASFA 120
121 NTSNDKIMYI DYANLTKVIV PGRFIYVDDG ILSFKVLQII DESNLRVQAV NSGYIASHKG 180
181 VNLPNTDVDL PPLSAKDMKD LQFGVRNGIH IVFASFIRTS EDVLSIRKAL GSEGQDIKII 240
241 SKIENQQGLD NFDEILEVTD GVMIARGDLG IEILAPEVLA IQKKLIAKCN LAGKPVICAT 300
301 QMLDSMTHNP RPTRAEVSDV GNAVLDGADC VMLSGETAKG DYPVNAVNIM AATALIAEST 360
361 IAHLALYDDL RDATPKPTST TETVAAAATA AILEQDGKAI VVLSTTGNTA RLLSKYRPSC 420
421 PIILVTRHAR TARIAHLYRG VFPFLYEPKR LDDWGEDVHR RLKFGVEMAR SFGMVDNGDT 480
481 VVSIQGFKGG VGHSNTLRIS TVGQEF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CDC19 PYK2 |
| View Details | Krogan NJ, et al. (2006) |
|
MSS51 PYK2 SSC1 |
| View Details | Gavin AC, et al. (2006) |

|
MRS6 PYK2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
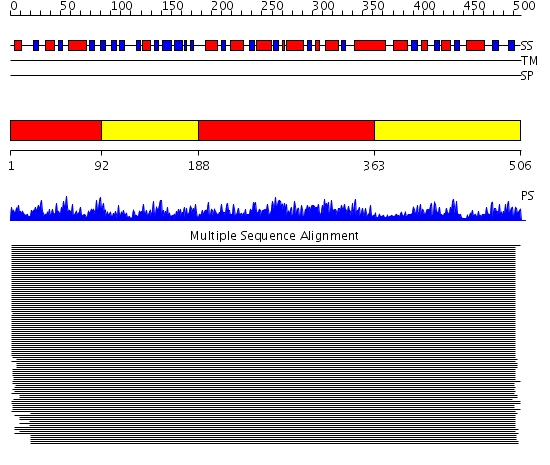
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] [188..362] |
11000.0 | Pyruvate kinase (PK); Pyruvate kinase, N-terminal domain; Pyruvate kinase, C-terminal domain | |
| 2 | View Details | [92..187] [363..506] |
11000.0 | Pyruvate kinase (PK); Pyruvate kinase, N-terminal domain; Pyruvate kinase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)