













| General Information: |
|
| Name(s) found: |
DYS1 /
YHR068W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 387 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
peptidyl-lysine modification to hypusine
[IDA |
| Molecular Function: |
transferase activity, transferring alkyl or aryl (other than methyl) groups
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDINEKLPE LLQDAVLKAS VPIPDDFVKV QGIDYSKPEA TNMRATDLIE AMKTMGFQAS 60
61 SVGTACEIID SMRSWRGKHI DELDDHEKKG CFDEEGYQKT TIFMGYTSNL ISSGVRETLR 120
121 YLVQHKMVDA VVTSAGGVEE DLIKCLAPTY LGEFALKGKS LRDQGMNRIG NLLVPNDNYC 180
181 KFEEWIVPIL DKMLEEQDEY VKKHGADCLE ANQDVDSPIW TPSKMIDRFG KEINDESSVL 240
241 YWAHKNKIPI FCPSLTDGSI GDMLFFHTFK ASPKQLRVDI VGDIRKINSM SMAAYRAGMI 300
301 ILGGGLIKHH IANACLMRNG ADYAVYINTG QEYDGSDAGA RPDEAVSWGK IKAEAKSVKL 360
361 FADVTTVLPL IVAATFASGK PIKKVKN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
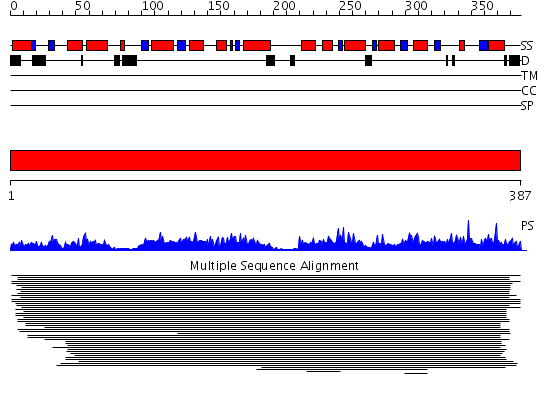
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..387] | 1340.0 | Deoxyhypusine synthase, DHS |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)