













| General Information: |
|
| Name(s) found: |
NUP188 /
YML103C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1655 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
mRNA export from nucleus
[TAS rRNA export from nucleus [TAS nuclear pore organization [IMP snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS protein export from nucleus [TAS ribosomal protein import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATPSFGNSS PQLTFTHVAN FMNDAAADVS AVDAKQLAQI RQFLKANKTN LIESLNTIRQ 60
61 NVTSSGDHNK LRSTIANLLQ INVDNDPFFA QSEDLSHAVE FFMSERSSRL HIVYSLLVNP 120
121 DIDLETYSFI DNDRFNVVGK LISIISSVIQ NYDIITASSL AHDYNNDQDM FTIVSLVQLK 180
181 KFSDLKFILQ ILQILNLMIL NTKVPVDIVN QWFLQYQNQF VEFCRNINST DKSIDTSSLQ 240
241 LYKFQNFQDL SYLSETLISR ISSLFTITTI LILGLNTSIA QFDIQSPLYM DTETFDTVNS 300
301 ALENDVATNI VNEDPIFHPM IHYSWSFILY YRRALQSSES FDDSDITKFA LFAESHDVLQ 360
361 KLNTLSEILS FDPVYTTVIT VFLEFSLNFI PITASTSRVF AKIISKAPEQ FIENFLTNDT 420
421 FEKKLSIIKA KLPLLNESLI PLINLALIDT EFANFELKDI CSFAVTKSSL NDLDYDLIAD 480
481 TITNSSSSSD IIVPDLIELK SDLLVAPPLE NENSNCLLSI PKSTKGKILT IKQQQQQQQQ 540
541 QNGQQPPTTS NLIIFLYKFN GWSLVGRILQ NLLHSYMEKG TQLDDLQHEL MISIIKLVTN 600
601 VVDPKTSIEK SSEILSYLSN SLDTSASTIN GASIIQVIFE IFEISLQRKD YTSIVQCCEF 660
661 MTMLTPNYLH LVSSYLNKSD LLDKYGKTGL SNMILGSVEL STGDYTFTIQ LLKLTKVFIR 720
721 ESLSLKNIHI SKRSKIDIIN KLILHAIHIF ESYYNWKYNN FLQKFEIAFH LTLIFYDVLH 780
781 DVFTINPHQK DQLIISSSAN KLLQLFLTPM DSIDLAPNTL TNILISPLNT TTKILGDKIL 840
841 GNLYSKVMNN SFKLCTLLIA IRGSNRDLKP SNLEKLLFIN SSKLVDVYTL PSYVHFKVQI 900
901 IELLSYLVEA PWNDDYPFLL SFLGEAKSMA FLKEVLSDLS SPVQDWNLLR SLYIFFTTLL 960
961 ESKQDGLSIL FLTGQFASNK KINDESSIDK KSSILTVLQK NSLLLDSTPE EVSCKLLETI1020
1021 TYVLNTWTNS KIFIKDPKFV NSLLAKLKDS KKLFQKKENL TRDETVSLIK KYKLISRIVE1080
1081 IFALCIYNST DSNSEILNFL NQEDLFELVH HFFQIDGFNK TFHDELNLKF KEKWPSLELQ1140
1141 SFQKIPLSRI NENENFGYDI PLLDIVLKAD RSWNEPSKSQ TNFKEEITDA SLNLQYVNYE1200
1201 ISTAKAWGAL ITTFVKRSTV PLNDGFVDLV EHFLKLNIDF GSDKQMFTQI YLERIELSFY1260
1261 ILYSFKLSGK LLKEEKIIEL MNKIFTIFKS GEIDFIKNIG KSLKNNFYRP LLRSVLVLLE1320
1321 LVSSGDRFIE LISDQLLEFF ELVFSKGVYL ILSEILCQIN KCSTRGLSTD HTTQIVNLED1380
1381 NTQDLLLLLS LFKKITNVNP SKNFNVILAS SLNEVGTLKV ILNLYSSAHL IRINDEPILG1440
1441 QITLTFISEL CSIEPIAAKL INSGLYSVLL ESPLSVAIQQ GDIKPEFSPR LHNIWSNGLL1500
1501 SIVLLLLSQF GIKVLPETCL FVSYFGKQIK STIYNWGDNK LAVSSSLIKE TNQLVLLQKM1560
1561 LNLLNYQELF IQPKNSDDQQ EAVELVIGLD SEHDKKRLSA ALSKFLTHPK YLNSRIIPTT1620
1621 LEEQQQLEDE SSRLEFVKGI SRDIKALQDS LFKDV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GLE2 NOP14 NUP1 NUP100 NUP116 NUP133 NUP157 NUP159 NUP170 NUP188 NUP2 NUP42 POM152 |
| View Details | Krogan NJ, et al. (2006) |
|
ABF2 ARG82 CKI1 CMR1 HHF1, HHF2 HTA1 HTA2 HTB1 HTB2 NUP188 PPH3 PSY2 PSY4 RRD1 SLX5 SLX8 SPT5 STP1 TOP2 |
| View Details | Qiu et al. (2008) |
|
ASM4 DBP5 GLE2 KAP120 MEX67 NDC1 NIC96 NSP1 NUP1 NUP100 NUP116 NUP120 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP192 NUP2 NUP42 NUP49 NUP53 NUP57 NUP60 NUP82 NUP84 NUP85 POM152 SEH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
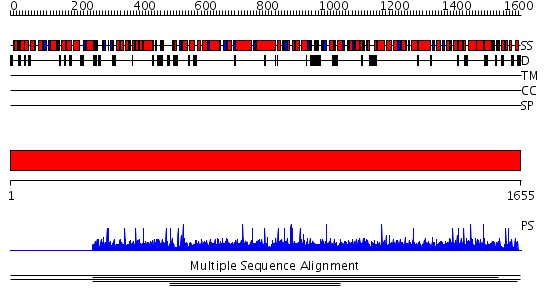
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1655] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [183..406] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [407..513] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [514..603] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [604..786] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [787..864] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [865..1050] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1051..1175] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1176..1378] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1379..1655] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)