













| General Information: |
|
| Name(s) found: |
NUP192 /
YJL039C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[TAS |
| Biological Process: |
nuclear pore organization
[IMP |
| Molecular Function: |
structural constituent of nuclear pore
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKWSAIPFQT LYRSIESGEF DFDLFKEVLP DLQNLNLNTD KLKNNASRSQ LEKGEIELSD 60
61 GSTFKVNQEF IFEAISLSDE LNLDEIVACE LILSGDTTAN NGKVQYFLRR QYILQIVSFI 120
121 VNCFHEDTEL YQELIKNGAL VSNILSAFKF IHTQLSEIKQ QINKAQILEN YNALFQQNIK 180
181 FRRDFLLREY DILSQILYGL VDKGAIMKNK DFILSLLHHV SELDSNDFFI IYYTPAFFHL 240
241 FASLRVLPDA DVKLLHSQFM KDLKDDSIYT KPVKVALIFI FFAYFIGWCK EDPKRRADTM 300
301 DFKTDVDEPM TSAVELGAIE QILIFAADTS IVEQDKSMEL FYDIRSLLER HIPRLIPKQL 360
361 LDDEKIFSQT TNSTYNPASA TDNMSGRGLW NPSYPGMMST TGTARLNSMP NNVNEYSYTT 420
421 IVLSDQTQEF FLSSFDDVLQ TIITDCAFLL TKIKDAEEDS LLSGEDLTLD DISLKADLER 480
481 FFLSIYFFYA SRPEYSCTFW SDKESNAYGF IEWCSRCNDN LMRSCFYLMV SSLSFGPENA 540
541 LNVYHYFGEN SSISWKNIAQ CLSDYTKKIS NFNSSLHKRQ QFSESTHNDI DSTAVALEEG 600
601 LNEEAVIFLS SLLTLVGSVT YQVDEDVKSS LSKVFSDVLF EFTKINTPLV GAAFKVISNL 660
661 VPKLESSRTK FWSFLDSLIF KDSSLNYSSE SYRNAFTNVL TKYSDVLGFL QLFHNLISIH 720
721 SRENNSEYMV FGKLAFPTRL GQGYRKVGIW PYFDYIFNDI LAHVDQIVDI RNKRAVQLPI 780
781 LKIIYTGLCS FDYSVILNSI PAAANLDALV DCENFFNYVQ ECPAIPIFNY IFTEKIYKSI 840
841 FNVVDVGVDQ LSIELEGGKN QAELLQLAVK IINKVLDYQE TYVEELFPIV KKHGKTDYFL 900
901 PKNYSLHGLR SFYDAIFFNI PLVAHLGLYV GVDDQILATN SLRILAKLSE RSNGSVASLS 960
961 KRNKLLTIFD SVDESARIKD AFITQLESSI TDAGVLALKL ELLDFLTSNL SNYSRTMTIS1020
1021 HLLLGFQVSN VISLGPNLAT FISSGTSLLD SLISVLEASL NSITKDNIDY APMRLATAAL1080
1081 EIILKLCRNP LTSGLLYSYL IKENFFERIM ILDPQVTRFT TWNGSPFDNS TEEKCKNFIE1140
1141 SESVGAFLSF LAYRNYWTQY LGLFIHKISF SGTKSEVLTY VNYLISNTMY SVRLFSFLDP1200
1201 LNYGNICEPK ETLSIFTNVP LNLEQVTLNK YCSGNIYDFH KMENLMRLIK RVRAESLHSN1260
1261 SFSLTVSKEQ FLKDADVECI KAKSHFTNII SRNKALELNL SVLHSWVQLV QIIVTDGKLE1320
1321 PSTRSNFILE VFGTIIPKIS DYIEFNITFS EELVSLAVFL FDIYNRDRKL ITDKGTVDGR1380
1381 LYQLFKTCIQ GINSPLSSVA LRSDFYILAN HYLSRVLSDQ VGSEKVLQDL RLGSKKLVEI1440
1441 IWNDVVYGEG TSRVTGILLL DSLIQLANRS KENFILDSLM KTTRLLLIIR SLKNTDALLN1500
1501 STTEHINIDD LLYELTAFKA TVFFLIRVAE TRGGASALIE NNLFRIIAEL SFLKVDPDLG1560
1561 LDLMFDEVYV QNSKFLKVNV TLDNPLLVDK DANGVSLFEL IVPIFQLISA VLVSMGSSNK1620
1621 AVVQTVKGLL NTYKRLVIGI FKRDLLREKE DKKNSSDPNN QSLNEMVKLI VMLCTLTGYQ1680
1681 NND |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
GDB1 NUP192 |
| View Details | Qiu et al. (2008) |
|
KAP120 NSP1 NUP1 NUP100 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP192 NUP53 NUP82 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
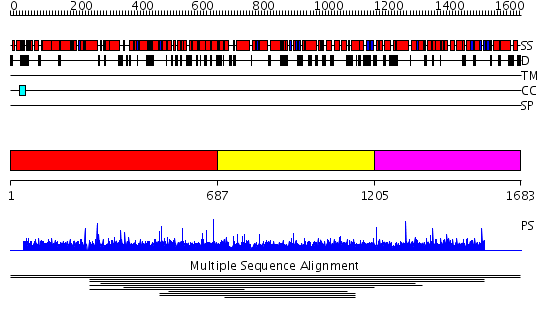
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..686] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [687..1204] | 1.009913 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1205..1683] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [564..684] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [685..852] | 1.051994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [853..961] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [962..1153] | 1.051971 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1154..1340] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1341..1449] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1450..1571] | 2.033961 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1572..1683] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)