













| General Information: |
|
| Name(s) found: |
PRP40 /
YKL012W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 583 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U1 snRNP
[IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IDA |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIWKEAKDA SGRIYYYNTL TKKSTWEKPK ELISQEELLL RENGWKAAKT ADGKVYYYNP 60
61 TTRETSWTIP AFEKKVEPIA EQKHDTVSHA QVNGNRIALT AGEKQEPGRT INEEESQYAN 120
121 NSKLLNVRRR TKEEAEKEFI TMLKENQVDS TWSFSRIISE LGTRDPRYWM VDDDPLWKKE 180
181 MFEKYLSNRS ADQLLKEHNE TSKFKEAFQK MLQNNSHIKY YTRWPTAKRL IADEPIYKHS 240
241 VVNEKTKRQT FQDYIDTLID TQKESKKKLK TQALKELREY LNGIITTSSS ETFITWQQLL 300
301 NHYVFDKSKR YMANRHFKVL THEDVLNEYL KIVNTIENDL QNKLNELRLR NYTRDRIARD 360
361 NFKSLLREVP IKIKANTRWS DIYPHIKSDP RFLHMLGRNG SSCLDLFLDF VDEQRMYIFA 420
421 QRSIAQQTLI DQNFEWNDAD SDEITKQNIE KVLENDRKFD KVDKEDISLI VDGLIKQRNE 480
481 KIQQKLQNER RILEQKKHYF WLLLQRTYTK TGKPKPSTWD LASKELGESL EYKALGDEDN 540
541 IRRQIFEDFK PESSAPTAES ATANLTLTAS KKRHLTPAVE LDY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
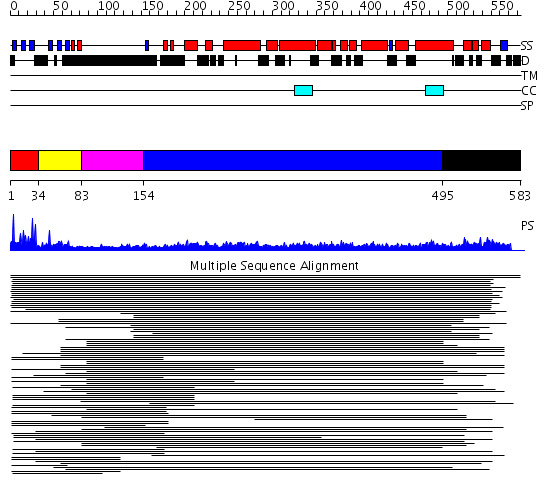
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..33] | 27.31133 | Formin binding protein FBP28 domain | |
| 2 | View Details | [34..82] | 8.30103 | Ubiquitin ligase NEDD4 WWIII domain | |
| 3 | View Details | [83..153] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [154..494] | 23.69897 | Heavy meromyosin subfragment | |
| 5 | View Details | [495..583] | 11.537602 | FF domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)