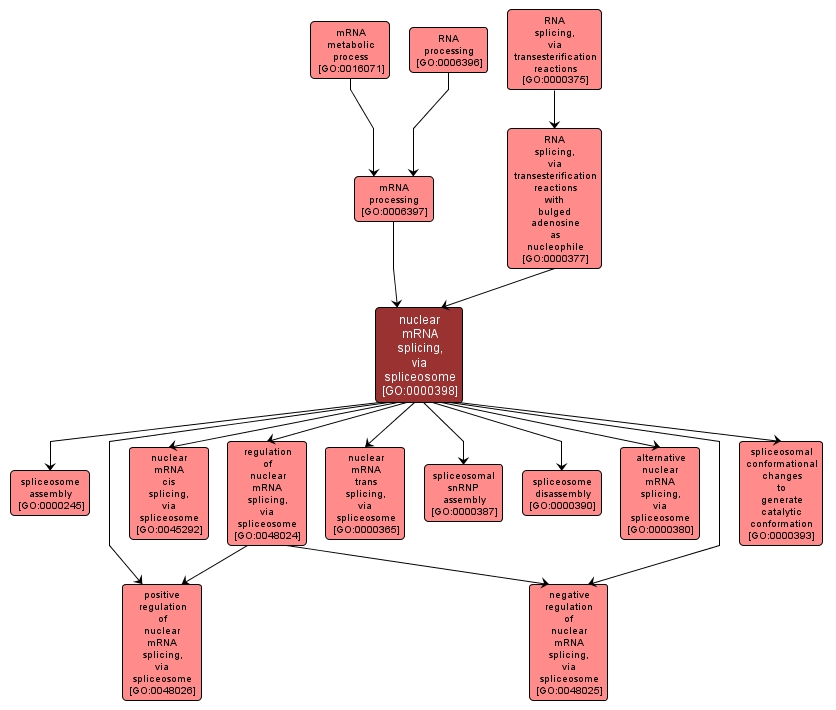
GO TERM SUMMARY
|
| Name: |
nuclear mRNA splicing, via spliceosome |
| Acc: |
GO:0000398 |
| Aspect: |
Biological Process |
| Desc: |
The joining together of exons from one or more primary transcripts of nuclear messenger RNA (mRNA) and the excision of intron sequences, via a spliceosomal mechanism, so that mRNA consisting only of the joined exons is produced. |
Synonyms:
- nuclear mRNA splicing via U2-type spliceosome
- mRNA splicing
- nuclear mRNA splicing via U12-type spliceosome
- splicing AT-AC intron
- GO:0006374
- splicing GT-AG intron
- GO:0006375
- pre-mRNA splicing
|
|

|
INTERACTIVE GO GRAPH
|