













| General Information: |
|
| Name(s) found: |
PRP24 /
YMR268C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 444 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U6 snRNP
[IDA |
| Biological Process: |
spliceosome assembly
[IMP assembly of spliceosomal tri-snRNP [IDA |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] U6 snRNA binding [IDA] RNA binding [IEA] snRNA binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEYGHHARPD SKRPLDEGSP AAAGLTSKKA NEALTRNREL TTVLVKNLPK SYNQNKVYKY 60
61 FKHCGPIIHV DVADSLKKNF RFARIEFARY DGALAAITKT HKVVGQNEII VSHLTECTLW 120
121 MTNFPPSYTQ RNIRDLLQDI NVVALSIRLP SLRFNTSRRF AYIDVTSKED ARYCVEKLNG 180
181 LKIEGYTLVT KVSNPLEKSK RTDSATLEGR EIMIRNLSTE LLDENLLRES FEGFGSIEKI 240
241 NIPAGQKEHS FNNCCAFMVF ENKDSAERAL QMNRSLLGNR EISVSLADKK PFLERNEVKR 300
301 LLASRNSKEL ETLICLFPLS DKVSPSLICQ FLQEEIHINE KDIRKILLVS DFNGAIIIFR 360
361 DSKFAAKMLM ILNGSQFQGK VIRSGTINDM KRYYNNQQNH SMKHVKPSCI NMMEKGPNLQ 420
421 VKKKIPDKQE QMSNDDFRKM FLGE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
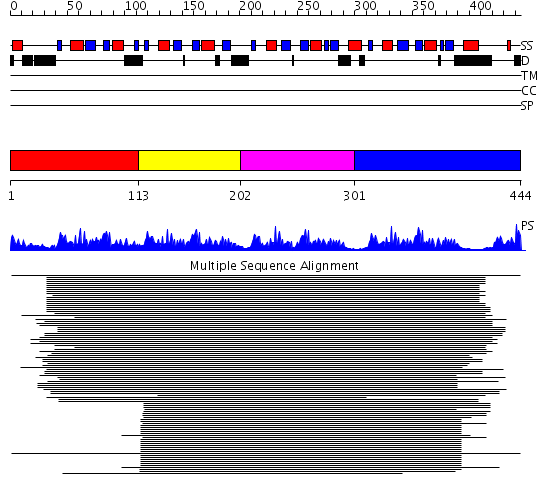
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | 54.69897 | Poly(A)-binding protein | |
| 2 | View Details | [113..201] | 57.0 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 3 | View Details | [202..300] | 57.0 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 4 | View Details | [301..444] | 16.30103 | Polypyrimidine tract-binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)