













| General Information: |
|
| Name(s) found: |
UTP21 /
YLR409C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 939 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear outer membrane [IPI nucleolus [IDA small nucleolar ribonucleoprotein complex [IPI |
| Biological Process: |
ribosome biogenesis
[RCA |
| Molecular Function: |
snoRNA binding
[RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIDLKKRKV EEDVRSRGKN SKIFSPFRII GNVSNGVPFA TGTLGSTFYI VTCVGKTFQI 60
61 YDANTLHLLF VSEKETPSSI VALSAHFHYV YAAYENKVGI YKRGIEEHLL ELETDANVEH 120
121 LCIFGDYLCA STDDNSIFIY KKSDPQDKYP SEFYTKLTVT EIQGGEIVSL QHLATYLNKL 180
181 TVVTKSNVLL FNVRTGKLVF TSNEFPDQIT TAEPAPVLDI IALGTVTGEV IMFNMRKGKR 240
241 IRTIKIPQSR ISSLSFRTDG SSHLSVGTSS GDLIFYDLDR RSRIHVLKNI HRESYGGVTQ 300
301 ATFLNGQPII VTSGGDNSLK EYVFDPSLSQ GSGDVVVQPP RYLRSRGGHS QPPSYIAFAD 360
361 SQSHFMLSAS KDRSLWSFSL RKDAQSQEMS QRLHKKQDGG RVGGSTIKSK FPEIVALAIE 420
421 NARIGEWENI ITAHKDEKFA RTWDMRNKRV GRWTFDTTDD GFVKSVAMSQ CGNFGFIGSS 480
481 NGSITIYNMQ SGILRKKYKL HKRAVTGISL DGMNRKMVSC GLDGIVGFYD FNKSTLLGKL 540
541 KLDAPITAMV YHRSSDLFAL ALDDLSIVVI DAVTQRVVRQ LWGHSNRITA FDFSPEGRWI 600
601 VSASLDSTIR TWDLPTGGCI DGIIVDNVAT NVKFSPNGDL LATTHVTGNG ICIWTNRAQF 660
661 KTVSTRTIDE SEFARMALPS TSVRGNDSML SGALESNGGE DLNDIDFNTY TSLEQIDKEL 720
721 LTLSIGPRSK MNTLLHLDVI RKRSKPKEAP KKSEKLPFFL QLSGEKVGDE ASVREGIAHE 780
781 TPEEIHRRDQ EAQKKLDAEE QMNKFKVTGR LGFESHFTKQ LREGSQSKDY SSLLATLINF 840
841 SPAAVDLEIR SLNSFEPFDE IVWFIDALTQ GLKSNKNFEL YETFMSLLFK AHGDVIHANN 900
901 KNQDIASALQ NWEDVHKKED RLDDLVKFCM GVAAFVTTA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
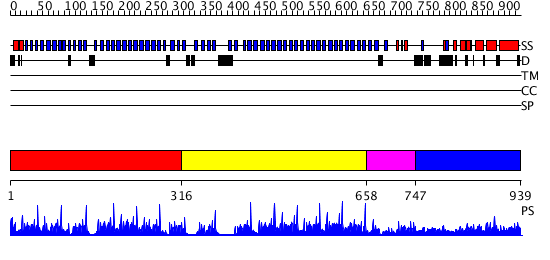
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..315] | 127.69897 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [316..657] | 110.9794 | Groucho/tle1, C-terminal domain | |
| 3 | View Details | [658..746] | 135.54902 | Tup1, C-terminal domain | |
| 4 | View Details | [747..939] | 1.118977 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)