













| General Information: |
|
| Name(s) found: |
PRP4 /
YPR178W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 465 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U4/U6 x U5 tri-snRNP complex
[IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKYIALENL PVDLQHKGAT QNESTADILK QLPHERLQAV LEKIPEEDLE VRRLLSILKK 60
61 PEVVENEDVQ QRRIRLAEIL MVDEIDLENI NNMENINGEE VDEEDDEDFF TPATSELIFA 120
121 RRFLINYSLE RSRKRLQKEM ERHQKFNTRQ ELLSRRTELQ RMANLELAGS QLVSTKPISA 180
181 VSLSTDDMVV ATGSWAGDLQ VLNSQTLQPL TQKLDSHVGK IGAIDWHPDS NNQMISCAED 240
241 GLIKNFQYSN EEGGLRLLGD LVGHERRISD VKYHPSGKFI GSASHDMTWR LWDASTHQEL 300
301 LLQEGHDKGV FSLSFQCDGS LVCSGGMDSL SMLWDIRSGS KVMTLAGHSK PIYTVAWSPN 360
361 GYQVATGGGD GIINVWDIRK RDEGQLNQIL AHRNIVTQVR FSKEDGGKKL VSCGYDNLIN 420
421 VYSSDTWLKM GSLAGHTDKI ISLDISNNSH FLVSGGWDRS IKLWN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
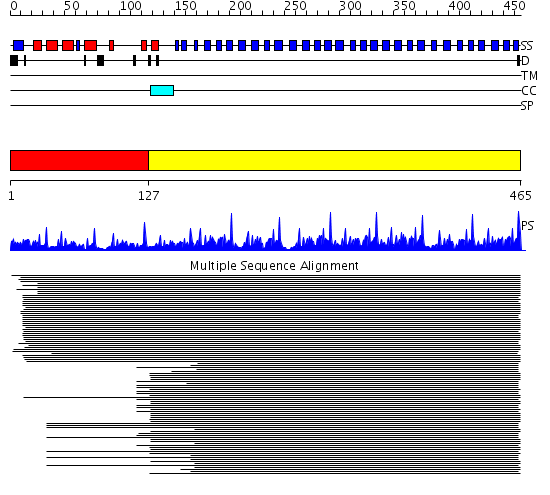
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 5.0 | Surface layer protein | |
| 2 | View Details | [127..465] | 339.450636 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)