













| General Information: |
|
| Name(s) found: |
GTS1 /
YGL181W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 396 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IPI |
| Biological Process: |
ascospore formation
[IMP drug transmembrane transport [IMP cell morphogenesis [IMP regulation of transcription [IMP ultradian rhythm [IMP chronological cell aging [IMP |
| Molecular Function: |
transcription coactivator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFRSSSHSL KHVDRELKEL INSSENANKC GECGNFYPTW CSVNLGVFLC GRCASVHRKV 60
61 FGSRDDDAFS NVKSLSMDRW TREDIDELVS LGGNKGNARF WNPKNVPFPF DGDDDKAIVE 120
121 HYIRDKYILG KFRYDEIKPE DFGSRMDDFD GESDRFDERN RSRSRSRSHS FYKGGHNRSD 180
181 YGGSRDSFQS SGSRYSRQLA ELKDMGFGDT NKNLDALSSA HGNINRAIDY LEKSSSSRNS 240
241 VSAAATTSTP PLPRRRATTS GPQPAIFDGT NVITPDFTSN SASFVQAKPA VFDGTLQQYY 300
301 DPATGMIYVD QQQYAMAMQQ QQQQQQQLAV AQAQAQAQAQ AQAQVQAQAQ AQAQAQAQAQ 360
361 QIQMQQLQMQ QQQQAPLSFQ QMSQGGNLPQ GYFYTQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BIM1 BMH1 GTS1 HAP1 RCN2 SFL1 |
| View Details | Qiu et al. (2008) |
|
CWC23 GTS1 PRP18 PRP2 PRP4 SPP2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
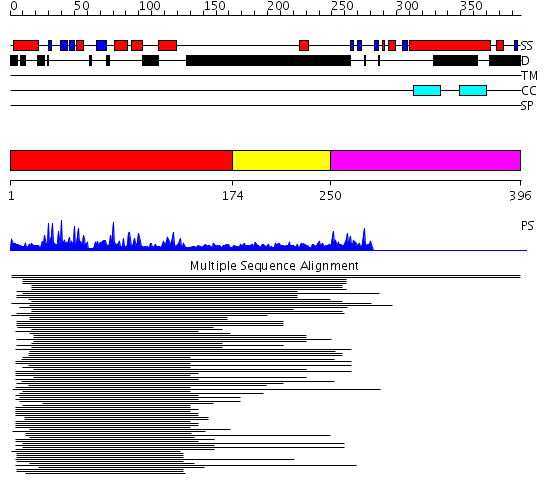
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 90.9794 | Pyk2-associated protein beta; Pyk2-associated protein beta ARF-GAP domain | |
| 2 | View Details | [174..249] | 5.29243 | UBA/TS-N domain No confident structure predictions are available. | |
| 3 | View Details | [250..396] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)