













| General Information: |
|
| Name(s) found: |
YAP1802 /
YGR241C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
actin cortical patch
[IDA |
| Biological Process: |
endocytosis
[IPI |
| Molecular Function: |
clathrin binding
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLYTKLVK GATKIKMAPP KQKYVDPILS GTSSARGLQE ITHALDIRLS DTAWTIVYKA 60
61 LIVLHLMIQQ GEKDVTLRHY SHNLDVFQLR KISHTTKWSS NDMRALQRYD EYLKTRCEEY 120
121 GRLGMDHLRD NYSSLKLGSK NQLSMDEELD HVESLEIQIN ALIRNKYSVS DLENHLLLYA 180
181 FQLLVQDLLG LYNALNEGVI TLLESFFELS IEHAKRTLDL YKDFVDMTEY VVRYLKIGKA 240
241 VGLKIPVIKH ITTKLINSLE EHLREETKRQ RGEPSEPQQD RKPSTAISST SSHNNNSNDK 300
301 NKSIAQKKLE QIREQKRLLE QQLQNQQLLI SPTVPQDAYN PFGSQQQDLN NDTFSFEPTQ 360
361 PQMTAQVPQP TANPFLIPQQ QQQALQLTSA STMPQPSEIQ ITPNLNNQQT GMYASNLQYT 420
421 PNFTGSGFGG YTTTENNAIM TGTLDPTKTG SNNPFSLENI AREQQQQNFQ NSPNPFTLQQ 480
481 AQTTPILAHS QTGNPFQAQN VVTSPMGTYM TNPVAGQLQY ASTGAQQQPQ MMQGQQTGYV 540
541 MVPTAFVPIN QQQQQQQHQQ ENPNLIDI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
ENT1 ENT2 PAN1 YAP1801 YAP1802 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
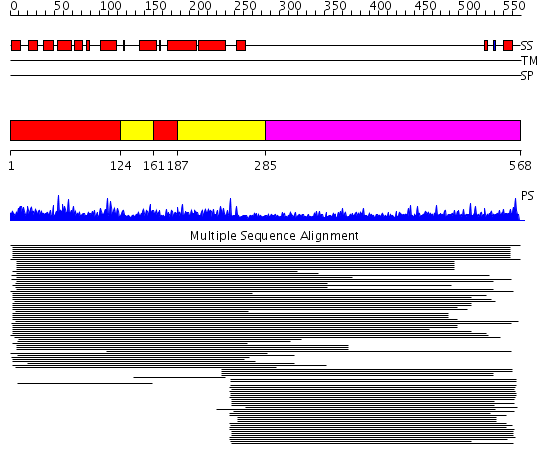
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] [161..186] |
325.0 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 2 | View Details | [124..160] [187..284] |
325.0 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 3 | View Details | [285..568] | 31.292997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)