













| General Information: |
|
| Name(s) found: |
GLN3 /
YER040W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 730 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
nitrogen compound metabolic process
[IPI positive regulation of transcription [IDA regulation of nitrogen utilization [IGI positive regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
transcription factor activity
[IDA transcription activator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQDDPENSKL YDLLNSHLDV HGRSNEEPRQ TGDSRSQSSG NTGENEEDIA FASGLNGGTF 60
61 DSMLEALPDD LYFTDFVSPF TAAATTSVTT KTVKDTTPAT NHMDDDIAMF DSLATTQPID 120
121 IAASNQQNGE IAQLWDFNVD QFNMTPSNSS GSATISAPNS FTSDIPQYNH GSLGNSVSKS 180
181 SLFPYNSSTS NSNINQPSIN NNSNTNAQSH HSFNIYKLQN NNSSSSAMNI TNNNNSNNSN 240
241 IQHPFLKKSD SIGLSSSNTT NSVRKNSLIK PMSSTSLANF KRAASVSSSI SNMEPSGQNK 300
301 KPLIQCFNCK TFKTPLWRRS PEGNTLCNAC GLFQKLHGTM RPLSLKSDVI KKRISKKRAK 360
361 QTDPNIAQNT PSAPATASTS VTTTNAKPIR SRKKSLQQNS LSRVIPEEII RDNIGNTNNI 420
421 LNVNRGGYNF NSVPSPVLMN SQSYNSSNAN FNGASNANLN SNNLMRHNSN TVTPNFRRSS 480
481 RRSSTSSNTS SSSKSSSRSV VPILPKPSPN SANSQQFNMN MNLMNTTNNV SAGNSVASSP 540
541 RIISSANFNS NSPLQQNLLS NSFQRQGMNI PRRKMSRNAS YSSSFMAASL QQLHEQQQVD 600
601 VNSNTNTNSN RQNWNSSNSV STNSRSSNFV SQKPNFDIFN TPVDSPSVSR PSSRKSHTSL 660
661 LSQQLQNSES NSFISNHKFN NRLSSDSTSP IKYEADVSAG GKISEDNSTK GSSKESSAIA 720
721 DELDWLKFGI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
GLN3 MET4 URE2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
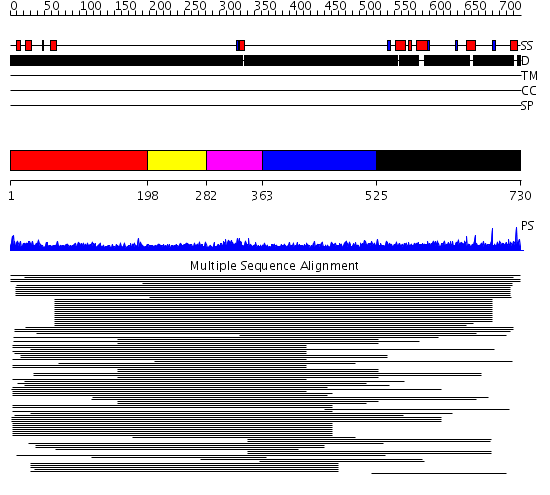
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 1.167996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [198..281] | 91.37734 | Erythroid transcription factor GATA-1 | |
| 3 | View Details | [282..362] | 149.383689 | Erythroid transcription factor GATA-1 | |
| 4 | View Details | [363..524] | 5.214996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [525..730] | 11.183996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)