













| General Information: |
|
| Name(s) found: |
HAP1 /
YLR256W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1502 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
aerobic respiration
[IMP positive regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
specific RNA polymerase II transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNTPYNSSV PSIASMTQSS VSRSPNMHTA TTPGANTSSN SPPLHMSSDS SKIKRKRNRI 60
61 PLSCTICRKR KVKCDKLRPH CQQCTKTGVA HLCHYMEQTW AEEAEKELLK DNELKKLRER 120
121 VKSLEKTLSK VHSSPSSNSL KSYNTPESSN LFMGSDEHTT LVNANTGSAS SASHMHQQQQ 180
181 QQQQQEQQQD FSRSANANAN SSSLSISNKY DNDELDLTKD FDLLHIKSNG TIHLGATHWL 240
241 SIMKGDPYLK LLWGHIFAMR EKLNEWYYQK NSYSKLKSSK CPINHAQAPP SAAAAATRKC 300
301 PVDHSAFSSG MVAPKEETPL PRKCPVDHTM FSSGMIPPRE DTSSQKRCPV DHTMYSAGMM 360
361 PPKDETPSPF STKAMIDHNK HTMNPPQSKC PVDHRNYMKD YPSDMANSSS NPASRCPIDH 420
421 SSMKNTAALP ASTHNTIPHH QPQSGSHARS HPAQSRKHDS YMTESEVLAT LCEMLPPKRV 480
481 IALFIEKFFK HLYPAIPILD EQNFKNHVNQ MLSLSSMNPT VNNFGMSMPS SSTLENQPIT 540
541 QINLPKLSDS CNLGILIIIL RLTWLSIPSN SCEVDLGEES GSFLVPNESS NMSASALTSM 600
601 AKEESLLLKH ETPVEALELC QKYLIKFDEL SSISNNNVNL TTVQFAIFYN FYMKSASNDL 660
661 TTLTNTNNTG MANPGHDSES HQILLSNITQ MAFSCGLHRD PDNFPQLNAT IPATSQDVSN 720
721 NGSKKANPST NPTLNNNMSA ATTNSSSRSG SADSRSGSNP VNKKENQVSI ERFKHTWRKI 780
781 WYYIVSMDVN QSLSLGSPRL LRNLRDFSDT KLPSASRIDY VRDIKELIIV KNFTLFFQID 840
841 LCIIAVLNHI LNVSLARSVR KFELDSLINL LKNLTYGTEN VNDVVSSLIN KGLLPTSEGG 900
901 SVDSNNDEIY GLPKLPDILN HGQHNQNLYA DGRNTSSSDI DKKLDLPHES TTRALFFSKH 960
961 MTIRMLLYLL NYILFTHYEP MGSEDPGTNI LAKEYAQEAL NFAMDGYRNC MIFFNNIRNT1020
1021 NSLFDYMNVI LSYPCLDIGH RSLQFIVCLI LRAKCGPLTG MRESSIITNG TSSGFNSSVE1080
1081 DEDVKVKQES SDELKKDDFM KDVNLDSGDS LAEILMSRML LFQKLTKQLS KKYNYAIRMN1140
1141 KSTGFFVSLL DTPSKKSDSK SGGSSFMLGN WKHPKVSNMS GFLAGDKDQL QKCPVYQDAL1200
1201 GFVSPTGANE GSAPMQGMSL QGSTARMGGT QLPPIRSYKP ITYTSSNLRR MNETGEAEAK1260
1261 RRRFNDGYID NNSNNDIPRG ISPKPSNGLS SVQPLLSSFS MNQLNGGTIP TVPSLTNITS1320
1321 QMGALPSLDR ITTNQINLPD PSRDEAFDNS IKQMTPMTSA FMNANTTIPS STLNGNMNMN1380
1381 GAGTANTDTS ANGSALSTLT SPQGSDLASN SATQYKPDLE DFLMQNSNFN GLMINPSSLV1440
1441 EVVGGYNDPN NLGRNDAVDF LPVDNVEIDG VGIKINYHLL TSIYVTSILS YTVLEDDAND1500
1501 EK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BIM1 BMH1 GTS1 HAP1 RCN2 SFL1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
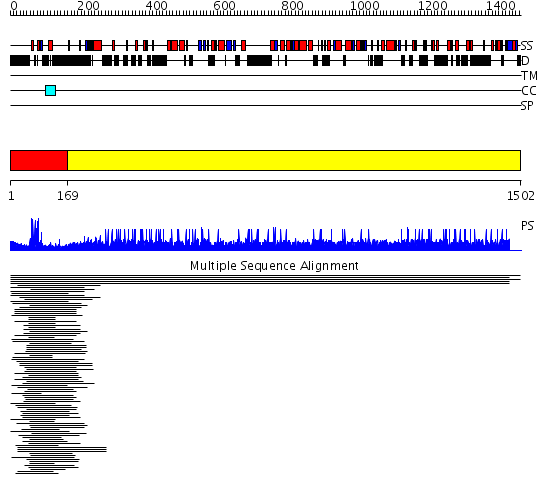
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 463.770868 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [169..1502] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [202..283] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [284..462] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [463..710] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [711..756] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [757..891] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [892..1073] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1074..1266] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1267..1387] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1388..1456] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1457..1502] | 1.002991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)