













| General Information: |
|
| Name(s) found: |
NOG1 /
YPL093W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[IMP nucleolus [IDA preribosome, large subunit precursor [IDA |
| Biological Process: |
ribosomal subunit export from nucleus
[IMP ribosomal large subunit biogenesis [IDA ribosome biogenesis [RCA |
| Molecular Function: |
GTP binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQLSWKDIPT VAPANDLLDI VLNRTQRKTP TVIRPGFKIT RIRAFYMRKV KYTGEGFVEK 60
61 FEDILKGFPN INDVHPFHRD LMDTLYEKNH YKISLAAISR AKSLVEQVAR DYVRLLKFGQ 120
121 SLFQCKQLKR AALGRMATIV KKLRDPLAYL EQVRQHIGRL PSIDPNTRTL LICGYPNVGK 180
181 SSFLRCITKS DVDVQPYAFT TKSLYVGHFD YKYLRFQAID TPGILDRPTE EMNNIEMQSI 240
241 YAIAHLRSCV LYFMDLSEQC GFTIEAQVKL FHSIKPLFAN KSVMVVINKT DIIRPEDLDE 300
301 ERAQLLESVK EVPGVEIMTS SCQLEENVME VRNKACEKLL ASRIENKLKS QSRINNVLNK 360
361 IHVAQPQARD DVKRTPFIPE SVKNLKKYDP EDPNRRKLAR DIEAENGGAG VFNVNLKDKY 420
421 LLEDDEWKND IMPEILDGKN VYDFLDPEIA AKLQALEEEE EKLENEGFYN SDDEEEIYDG 480
481 FEASEVDDIK EKAAWIRNRQ KTMIAEARNR KSLKNKAIMP RSKLTKSFGK MEEHMSTLGH 540
541 DMSALQDKQN RAARKNRYVE RGSDVVFGDQ DALTASTENG VKLRQTDRLL DGVADGSMRS 600
601 KADRMAKMER RERNRHAKQG ESDRHNAVSL SKHLFSGKRG VGKTDFR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | control 2 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
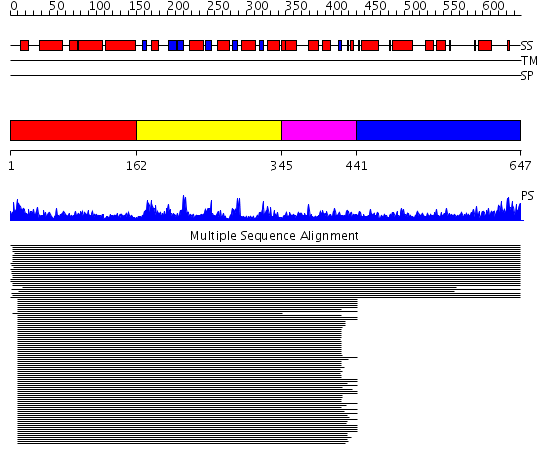
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 64.09691 | Probable GTPase Der, N-terminal and middle domains; Probable GTPase Der, C-terminal domain | |
| 2 | View Details | [162..344] | 64.09691 | Probable GTPase Der, N-terminal and middle domains; Probable GTPase Der, C-terminal domain | |
| 3 | View Details | [345..440] | 64.09691 | Probable GTPase Der, N-terminal and middle domains; Probable GTPase Der, C-terminal domain | |
| 4 | View Details | [441..647] | 2.205961 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)