













| General Information: |
|
| Name(s) found: |
AFG2 /
YLR397C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 780 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IDA |
| Biological Process: |
response to drug
[IMP |
| Molecular Function: |
ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPKSSSSGS KKKSSASSNS ADAKASKFKL PAEFITRPHP SKDHGKETCT AYIHPNVLSS 60
61 LEINPGSFCT VGKIGENGIL VIARAGDEEV HPVNVITLST TIRSVGNLIL GDRLELKKAQ 120
121 VQPPYATKVT VGSLQGYNIL ECMEEKVIQK LLDDSGVIMP GMIFQNLKTK AGDESIDVVI 180
181 TDASDDSLPD VSQLDLNMDD MYGGLDNLFY LSPPFIFRKG STHITFSKET QANRKYNLPE 240
241 PLSYAAVGGL DKEIESLKSA IEIPLHQPTL FSSFGVSPPR GILLHGPPGT GKTMLLRVVA 300
301 NTSNAHVLTI NGPSIVSKYL GETEAALRDI FNEARKYQPS IIFIDEIDSI APNRANDDSG 360
361 EVESRVVATL LTLMDGMGAA GKVVVIAATN RPNSVDPALR RPGRFDQEVE IGIPDVDARF 420
421 DILTKQFSRM SSDRHVLDSE AIKYIASKTH GYVGADLTAL CRESVMKTIQ RGLGTDANID 480
481 KFSLKVTLKD VESAMVDIRP SAMREIFLEM PKVYWSDIGG QEELKTKMKE MIQLPLEASE 540
541 TFARLGISAP KGVLLYGPPG CSKTLTAKAL ATESGINFLA VKGPEIFNKY VGESERAIRE 600
601 IFRKARSAAP SIIFFDEIDA LSPDRDGSST SAANHVLTSL LNEIDGVEEL KGVVIVAATN 660
661 RPDEIDAALL RPGRLDRHIY VGPPDVNARL EILKKCTKKF NTEESGVDLH ELADRTEGYS 720
721 GAEVVLLCQE AGLAAIMEDL DVAKVELRHF EKAFKGIARG ITPEMLSYYE EFALRSGSSS 780
781 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |
|
AFG2 TDH3 YAR044W |
| View Details | Ho Y, et al. (2002) |
|
AFG2 ALB1 ARX1 BUD20 DSS4 EHD3 FAA4 FYV4 HHF1, HHF2 HTA1 HTB1 HTB2 LHP1 MAM33 MNP1 MRPS16 NMD3 NOG1 NOP12 NOP13 NOP7 NSA2 NUG1 PRP43 PUF6 PWP1 RIX1 RSM24 RSM25 SEC4 YBL044W YPT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
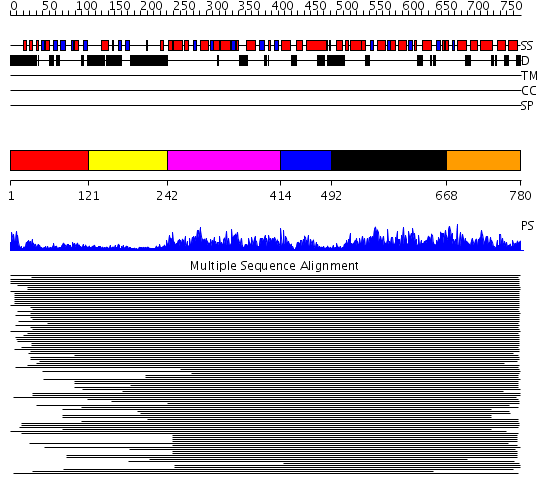
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 766.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [121..241] | 766.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [242..413] | 766.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [414..491] | 766.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [492..667] | 71.045757 | Hexamerization domain of N-ethylmalemide-sensitive fusion (NSF) protein | |
| 6 | View Details | [668..780] | 14.9 | AAA domain of cell division protein FtsH |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)