













| General Information: |
|
| Name(s) found: |
SPB1 /
YCL054W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 841 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
rRNA methylation
[IDA processing of 27S pre-rRNA [IDA |
| Molecular Function: |
rRNA (uridine-2'-O-)-methyltransferase activity
[IGI rRNA (guanine) methyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKTQKKNSK GRLDRYYYLA KEKGYRARSS FKIIQINEKY GHFLEKSKVV IDLCAAPGSW 60
61 CQVASKLCPV NSLIIGVDIV PMKPMPNVIT FQSDITTEDC RSKLRGYMKT WKADTVLHDG 120
121 APNVGLGWVQ DAFTQSQLTL QALKLAVENL VVNGTFVTKI FRSKDYNKLI WVFQQLFEKV 180
181 EATKPPASRN VSAEIFVVCK GFKAPKRLDP RLLDPKEVFE ELPDGQQNME SKIYNPEKKV 240
241 RKRQGYEEGD NLLYHETSIL DFVRTEDPIS MLGEMNKFTI DENDHEWKIL KKLKQTTDEF 300
301 RSCIEDLKVL GKKDFKMILR WRKIAREILG IEVKDDAKTE IEVVPLTEEE QIEKDLQGLQ 360
361 EKQRLNVKRE RRRKNEMKQK ELQRMQMNMI TPTDIGIEAA SLGKESLFNL KTAEKTGILN 420
421 DLAKGKKRMI FTDDELAKDN DIYIDENIMI KDKDSAADAD DLESELNAMY SDYKTRRSER 480
481 DAKFRAKQAR GGDNEEEWTG FNEGSLEKKE EEGKDYIEDN DDEGVEGDSD DDEAITNLIS 540
541 KLKGQEGDHK LSSKARMIFN DPIFNNVEPD LPVNTVNDGI MSSESVGDIS KLNKKRKHEE 600
601 MHQKQDEADS SDESSSDDSD FEIVANDNAS EEFDSDYDSE EEKNQTKKEK HSRDIDIATV 660
661 EAMTLAHQLA LGQKNKHDLV DEGFNRYTFR DTENLPDWFL EDEKEHSKIN KPITKEAAMA 720
721 IKEKIKAMNA RPIKKVAEAK ARKRMRAVAR LEKIKKKAGL INDDSDKTEK DKAEEISRLM 780
781 RKVTKKPKTK PKVTLVVASG RNKGLAGRPK GVKGKYKMVD GVMKNEQRAL RRIAKKHHKK 840
841 K |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
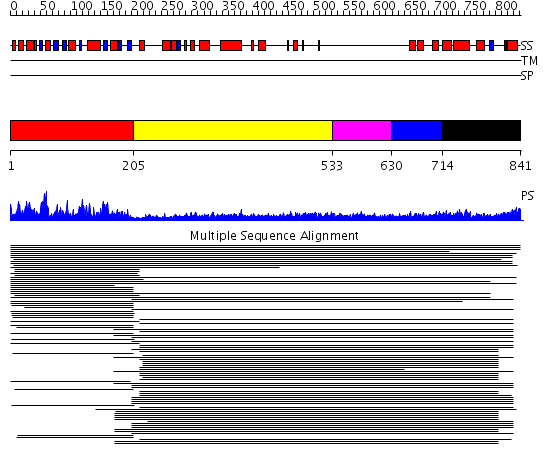
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..204] | 226.679424 | RNA methyltransferase FtsJ | |
| 2 | View Details | [205..532] | 16.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [533..629] | 5.0 | Tropomyosin | |
| 4 | View Details | [630..713] | 5.0 | Tropomyosin | |
| 5 | View Details | [714..841] | 8.703997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)