













| General Information: |
|
| Name(s) found: |
MHP1 /
YJL042W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1398 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule
[IDA |
| Biological Process: |
cellular cell wall organization
[IMP negative regulation of microtubule depolymerization [IMP |
| Molecular Function: |
structural constituent of cytoskeleton
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSKDTQKLL KEHRIPCIDV GWLVRPSAST SKSSRPGKSE SKANSVAPDI QMDTARPPVF 60
61 ETSVDSSSSI LSSNDKGRRH SVAASLLMDN QRANAGSTSV PTNIPPPRGR SKSVVETNLS 120
121 NVEADSGHHH HHRHHHHTED APAPKKVGFF KSLFGHRKKD QEQQEKERER KERSPSPTHV 180
181 DRGAAIRRER TATISAESPP PLQYNAPPSY NDTVVPLTRS KTESEVYYEN HPQSYYHGRM 240
241 RTYHSPEEGK VDGTSPADDH NYGGSRPDPR LMDFLRYYKS KDYKLAAFKE GNFIKSSASP 300
301 TTKKNRRASF SLHNDKPQPA KSLAHQKFDA KGRPIPPHPD APKLPSAFRK KHPSNASIVD 360
361 TVDSNSDVSS SAQNNNQTPS SHKFGAFLRK VTSYGNNNNN STNASSLSAN VNNPDTSSTS 420
421 LWSSSSMEFD PSKITTVPGL ENIRPLKHVS FATNTYFNDP PQQICSKNPR KGEVEVKPNG 480
481 SVVIHRLTPQ ERKKIMESTS LGVVVGGTGQ LKLLNPEEDD ANAKSKEEMA PQKQNEVEAH 540
541 DEEDNNSQRR NIVMAAAEAA AEARAKEAPN ELKRIVTNNE EEVTVSKTAS HLTIDKPMIS 600
601 RRGASTSSLA SMVSSDTNGT NADDEGEILP PPSLKIPHDI VYTRCCHLRE ILPIPATLKQ 660
661 LKKGSTDPIP ILQLRNPRPS MVEIWSFSDF LSIAPVLCLS LDGVQLTVQM LRIILSSLVY 720
721 KQHFQKLSLR NTPLDEEGWK VLCYFVSKAK SLHSIDLTMV PSIKTNVQKP SKSSLKSKIL 780
781 RMQCNLENRS DMNWDLLTAS IALMGGLEEI VISGAKMNSA QFKNFILVAC IATERLGLAY 840
841 NGLSKSQCDD LAKWMVQSKV TGLDVGFNDL NGKLSSFTDA VLGKIQKANE KNVFKFLSLN 900
901 GTNLRVNEHD TFENNEVLKL ISVLCYLENL KFLDISNNPA IFPHCVPTLI DFLPVFVNLV 960
961 RLHIDYNNLS STSVVMLAEI LPMCSRLNYF SMLGTELDLA SSKALAEAVR KSSSLMTLDV1020
1021 DYVYMPENIK EKISLYALRN IQGELKRVNS DDKDIKDSQF SSLQDQLSLL LTEKADNSEH1080
1081 YNKMVENFMA KIALARIKIS KVVHDLFDLK LNGQLNLEGK EALIRLCFIE ASLERGCDLL1140
1141 KQRHNNTLKS PEAVSKSRKG GNQAQPNSES CQRMLLSSSI LQNSDHIALM PFGSAIVEKS1200
1201 SPDAEDAVEF REGDDSNVNH EDVPANDQQF RDEVDIKNKY SIIKRELEHE KLVGGGDLPV1260
1261 DKEILNRAAQ SLDSDQIKEF LLKNDVSTIL GVIDELHSQG YHLHHIFKKQ GNQEETAFRT1320
1321 KDEQQSSQSN DSSANASPTT DPISTGSNTS RTNDNAHIPP TDAPGFDKFM NNAEENAIDA1380
1381 AYDDVLDKIQ DARNSSTK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ASE1 ATG4 ATG8 BIK1 BIM1 CBF5 CIN2 CIN4 MHP1 RBL2 SPC98 STU1 STU2 YTM1 |
| View Details | Ho Y, et al. (2002) |

|
CFT1 CIC1 CLU1 ERB1 FIN1 FIP1 FPR3 FPR4 GLC7 GLC8 GPH1 HER1 KEL1 MHP1 NSA1 REG1 RRP17 SCD5 SDS22 SEN1 SPB1 SUI2 SUI3 YTM1 |
| View Details | Qiu et al. (2008) |
|
ARC40 BIM1 CNM67 DYN1 HSK3 KIP2 KIP3 MHP1 SPC19 SPC34 STU1 TUB1 TUB3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
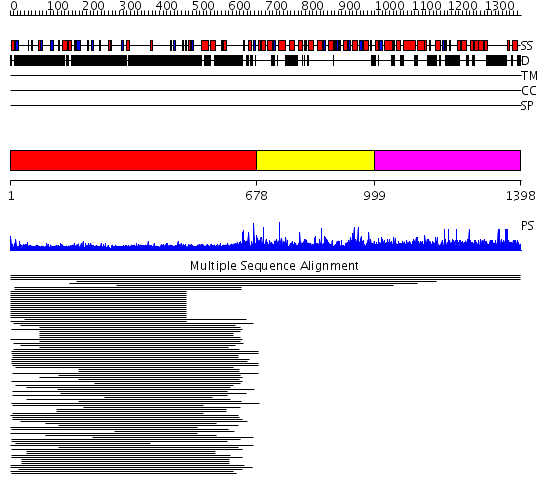
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..677] | 65.409998 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [678..998] | 2.009997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [999..1398] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [414..1185] | 2.41 | The molecular structure of toll-like receptor 3 ligand binding domain | |
| 5 | View Details | [1186..1398] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)