













| General Information: |
|
| Name(s) found: |
STU1 /
YBL034C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1513 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spindle pole body
[IDA |
| Biological Process: |
microtubule nucleation
[IPI |
| Molecular Function: |
structural constituent of cytoskeleton
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSFNNETNN NSNTNTHPDD SFPLYTVFKD ESVPIEEKMA LLTRFKGHVK KELVNESSIQ 60
61 AYFAALLFIS GHYAYRSYPR LIFLSHSSLC YLIKRVAMQS PVQFNDTLVE QLLNHLIFEL 120
121 PNEKKFWLAS IKAIEAIYLV NPSKIQAILA NFLRRPSENQ NGDYLNRIKS TLLTIDELIQ 180
181 INEKNNSNHL QLLRFFMLSF TNLLNNNLNE HANDDNNNVI IELIFDIMYK YLKMDDENSQ 240
241 DLIDGFINDL EVEKFKQKFI SLAKSQDQHG SQEDKSTLFD EEYEFQLLLA EAKLPQLSNN 300
301 LSSKDPAMKK NYESLNQLQQ DLENLLAPFQ SVKETEQNWK LRQSNIIELD NIISGNIPKD 360
361 NPEEFVTVIK EVQLIELISR ATSSLRTTLS LTALLFLKRL IHILNDQLPL SILDQIFVIF 420
421 KNLLSSTKKI SSQTAFHCLI TLIIDINHFH NKLFQLSFLL INEKTVTPRF CSAILLRSFL 480
481 IKFNDSNLSL NNSNTTSPTS KLENNIIYIE EWLKKGISDS QTTVREAMRL TFWYFYKCYP 540
541 TNAKRLLSSS FSPQLKKATE LAIPAHLNIN YQVSRVSSTA SASSATSRLY SHSSNNSSRK 600
601 TSLLEQKRNY PSYAQPTQSS STSLLNAPAV TAGGSVIASK LSNKLKTNLR STSEYSSKEN 660
661 EKRARHHDSM NSVSNSNTKD NNNVTKRKVS APPSSTAATK VSENYTNFDD FPSNQIDLTD 720
721 ELSNSYSNPL IKKYMDKNDV SMSSSPISLK GSNKLGEYET LYKKFNDASF PAQIKDALQY 780
781 LQKELLLTSQ QSSSAPKFEF PMIMKKLRQI MIKSPNDFKP FLSIEKFTNG VPLNYLIELY 840
841 SINSFDYAEI LKNRMNPEKP YELTNLIITI ADLFNFLNAN NCPNDFKLYY MKYKTTFFNY 900
901 NFKLLLEIFR NLNIKHDNTL RSGTNDLMPK ISMILFQIYG KEFDYTCYFN LIFEIYKFDN 960
961 NRFNKLLADF DIVSTKMKIC HELEKKDANF KVEDIISRES SVSFTPIDNK KSEGDEESDD1020
1021 AVDENDVKKC MEMTMINPFK NLETDKTLEL KNNVGKRTSS TDSVVIHDDN DKDKKLSEMT1080
1081 KIVSVYQLDQ PNPAKEEDDI DMENSQKSDL NLSEIFQNSG ENTERKLKDD NEPTVKFSTD1140
1141 PPKIINEPEK LIGNGNENEK PDLETMSPIK INGDENMGQK QRITVKRERD VALTEQDINS1200
1201 KKMKLVNNKK SEKMHLLIMD NFPRDSLTVY EISHLLMVDS NGNTLMDFDV YFNHMSKAIN1260
1261 RIKSGSFTMK HINYLIEPLI TCFQNQKMTD WLTNENGFDE LLDVAIMLLK STDDTPSIPS1320
1321 KISSKSIILV HCLLVWKKFL NTLSENADDD GVSVRMCFEE VWEQILLMLN KFSDYGNEIY1380
1381 KLAQEFRDSL MLSHFFKKHS ATRILSMLVT EIQPDTAGVK ETFLIETLWK MLQSPTICQQ1440
1441 FKKSNISEII QTMSYFIMGT DNTSWNFTSA VVLARCLRVL QTTPDYTEQE TERLFDCLPK1500
1501 NVFKMIMFIA SNE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
AME1 ARP1 ASE1 ATG4 ATG8 BIK1 BIM1 CBF5 CIK1 CIN2 CIN4 DYN1 JNM1 KAR3 KAR5 MHP1 NUF2 RBL2 SPC19 SPC34 SPC98 STU1 STU2 TID3 YTM1 |
| View Details | Ho Y, et al. (2002) |
|
ADR1 BMH1 BNR1 BOI2 CSR2 CYK3 GSY2 KCS1 NTH1 REG1 SOK1 STU1 SVL3 YFR017C YIL028W |
| View Details | Qiu et al. (2008) |

|
ARP1 ASE1 BBP1 BFA1 BIK1 BIM1 BUB1 CDC31 CIK1 CIN8 CNM67 DYN1 JNM1 KAR3 KAR9 KIP1 KIP2 MHP1 NDC1 NUD1 NUF2 SGO1 SLK19 SPC105 SPC110 SPC19 SPC24 SPC25 SPC29 SPC34 SPC42 SPC72 SPC97 SPC98 STU1 STU2 TID3 TUB2 TUB4 VIK1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3906: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
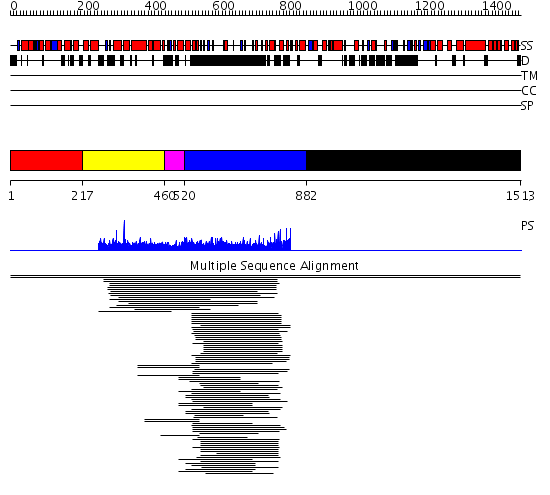
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [217..459] | 2.038996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [460..519] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [520..881] | 270.841998 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [882..1513] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1318..1513] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)