













| General Information: |
|
| Name(s) found: |
CIT2 /
YCR005C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 460 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA mitochondrion [IDA |
| Biological Process: |
glutamate biosynthetic process
[TAS glyoxylate cycle [TAS citrate metabolic process [TAS |
| Molecular Function: |
citrate (Si)-synthase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVPYLNSNR NVASYLQSNS SQEKTLKERF SEIYPIHAQD VRQFVKEHGK TKISDVLLEQ 60
61 VYGGMRGIPG SVWEGSVLDP EDGIRFRGRT IADIQKDLPK AKGSSQPLPE ALFWLLLTGE 120
121 VPTQAQVENL SADLMSRSEL PSHVVQLLDN LPKDLHPMAQ FSIAVTALES ESKFAKAYAQ 180
181 GISKQDYWSY TFEDSLDLLG KLPVIAAKIY RNVFKDGKMG EVDPNADYAK NLVNLIGSKD 240
241 EDFVDLMRLY LTIHSDHEGG NVSAHTSHLV GSALSSPYLS LASGLNGLAG PLHGRANQEV 300
301 LEWLFALKEE VNDDYSKDTI EKYLWDTLNS GRVIPGYGHA VLRKTDPRYM AQRKFAMDHF 360
361 PDYELFKLVS SIYEVAPGVL TEHGKTKNPW PNVDAHSGVL LQYYGLKESS FYTVLFGVSR 420
421 AFGILAQLIT DRAIGASIER PKSYSTEKYK ELVKNIESKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CIT2 PEP3 PRO2 PTC6 |
| View Details | Qiu et al. (2008) |
|
CIT1 CIT2 CIT3 MDH3 MLS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
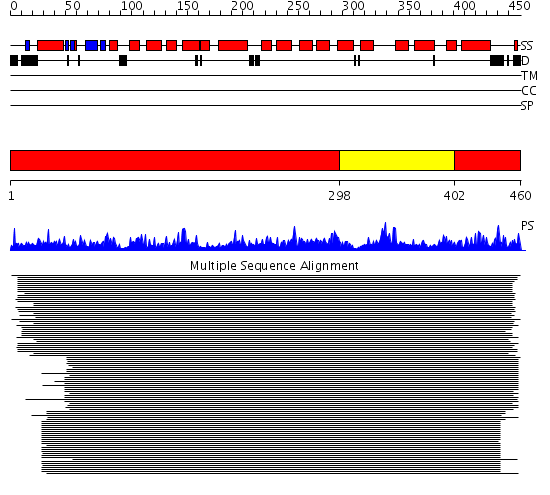
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] [402..460] |
1760.0 | Citrate synthase | |
| 2 | View Details | [298..401] | 1760.0 | Citrate synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)