













| General Information: |
|
| Name(s) found: |
BFR1 /
YOR198C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polysome
[IDA ribonucleoprotein complex [IDA nuclear membrane-endoplasmic reticulum network [IDA |
| Biological Process: |
mRNA metabolic process
[IPI regulation of mitosis [IMP meiosis [IMP |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSQQHKFKR PDVSVRDKKL DTLNVQLKKI DTEIGLIRKQ IDQHQVNDTT QQERKKLQDK 60
61 NKEIIKIQAD LKTRRSNIHD SIKQLDAQIK RKNNQIEEKL GKKAKFSSTA EAKQRINEIE 120
121 ESIASGDLSL VQEKLLVKEM QSLNKLIKDL VNIEPIRKSV DADKAKINQL KEELNGLNPK 180
181 DVSNQFEENQ QKLNDIHSKT QGVYDKRQTL FNKRAALYKK RDELYSQIRQ IRADFDNEFK 240
241 SFRAKLDKER LKREEEQRLS KLLEQKDVDM GKLQEKLTHA KIPAFTYEIG AIENSLLVLD 300
301 PTYVKPKKNI LPDLSSNALE TKPARKVVAD DLVLVTPKKD DFVNVAPSKS KKYKKKNQQK 360
361 NTENEQPASI FNKVDGKFTL EPTLIATLAE LDVTVPINSD DVKITVEQLK KKHEELLSKQ 420
421 EEQTKQNIES VEKEIEKLNL DYSNKEQQVK KELEEKRLKE QEESEKDKEN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
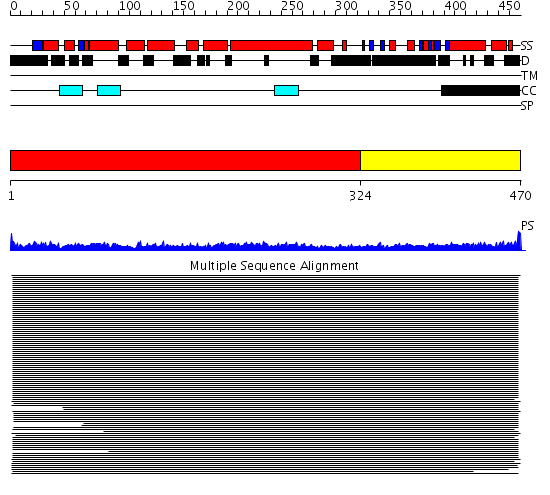
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..323] | 19.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [324..470] | 4.078997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)