













| General Information: |
|
| Name(s) found: |
SSZ1 /
YHR064C
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 538 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
regulation of translational fidelity
[IMP translation [IGI |
| Molecular Function: |
unfolded protein binding
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSPVIGITF GNTSSSIAYI NPKNDVDVIA NPDGERAIPS ALSYVGEDEY HGGQALQQLI 60
61 RNPKNTIINF RDFIGLPFDK CDVSKCANGA PAVEVDGKVG FVISRGEGKE EKLTVDEVVS 120
121 RHLNRLKLAA EDYIGSAVKE AVLTVPTNFS EEQKTALKAS AAKIGLQIVQ FINEPSAALL 180
181 AHAEQFPFEK DVNVVVADFG GIRSDAAVIA VRNGIFTILA TAHDLSLGGD NLDTELVEYF 240
241 ASEFQKKYQA NPRKNARSLA KLKANSSITK KTLSNATSAT ISIDSLADGF DYHASINRMR 300
301 YELVANKVFA QFSSFVDSVI AKAELDPLDI DAVLLTGGVS FTPKLTTNLE YTLPESVEIL 360
361 GPQNKNASNN PNELAASGAA LQARLISDYD ADELAEALQP VIVNTPHLKK PIGLIGAKGE 420
421 FHPVLLAETS FPVQKKLTLK QAKGDFLIGV YEGDHHIEEK TLEPIPKEEN AEEDDESEWS 480
481 DDEPEVVREK LYTLGTKLME LGIKNANGVE IIFNINKDGA LRVTARDLKT GNAVKGEL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
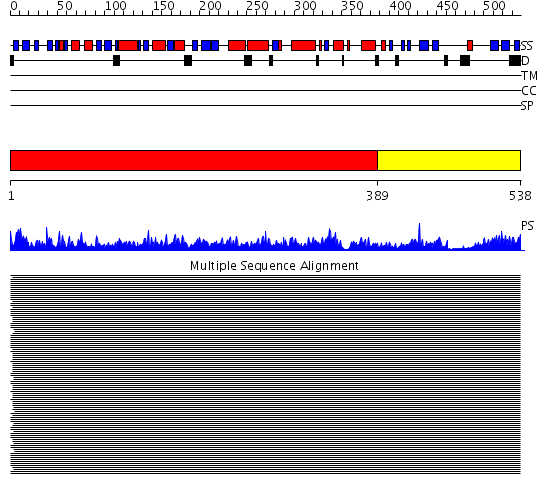
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..388] | 176.0 | Heat shock protein 70kDa, ATPase fragment | |
| 2 | View Details | [389..538] | 41.69897 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)