













| General Information: |
|
| Name(s) found: |
HFM1 /
YGL251C
[SGD]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1187 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
synapsis
[IGI DNA unwinding involved in replication [IDA meiosis [IDA |
| Molecular Function: |
DNA helicase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTKFDRLGT GKRSRPSPNN IDFNDQSATF KRNKKNSRQP SFKVGLSYNS LLDDCDDENE 60
61 TEEIFEGRGL QFFDKDDNFS ITADDTQVTS KLFDHDLEQT PDEEAKKPKK VTIRKSAKKC 120
121 LSTTILPDSF RGVFKFTEFN KMQSEAFPSI YESNENCIIS SPTGSGKTVL FELAILRLIK 180
181 ETNSDTNNTK IIYIAPTKSL CYEMYKNWFP SFVNLSVGML TSDTSFLETE KAKKCNIIIT 240
241 TPEKWDLLTR RWSDYSRLFE LVKLVLVDEI HTIKEKRGAS LEVILTRMNT MCQNIRFVAL 300
301 SATVPNIEDL ALWLKTNNEL PANILSFDES YRQVQLTKFV YGYSFNCKND FQKDAIYNSK 360
361 LIEIIEKHAD NRPVLIFCPT RASTISTAKF LLNNHIFSKS KKRCNHNPSD KILNECMQQG 420
421 IAFHHAGISL EDRTAVEKEF LAGSINILCS TSTLAVGVNL PAYLVIIKGT KSWNSSEIQE 480
481 YSDLDVLQMI GRAGRPQFET HGCAVIMTDS KMKQTYENLI HGTDVLESSL HLNLIEHLAA 540
541 ETSLETVYSI ETAVNWLRNT FFYVRFGKNP AAYQEVNRYV SFHSVEDSQI NQFCQYLLDT 600
601 LVKVKIIDIS NGEYKSTAYG NAMTRHYISF ESMKQFINAK KFLSLQGILN LLATSEEFSV 660
661 MRVRHNEKKL FKEINLSPLL KYPFLTEKKQ SQIIDRVSQK VSLLIQYELG GLEFPSYEGA 720
721 SKLHQTLVQD KFLVFRHCFR LLKCMVDTFI EKSDGTSLKN TLFLLRSLNG HCWENTPMVL 780
781 RQLKTIGLVS VRRLIRHGIT NLEEMGHLSD TQIEYYLNLK IGNGIKIKND ISLLPCLNIR 840
841 TKLENCKIEN EELWLTFKVE ISATFKSSIW HGQHLSLDIE TEKSSGELID FRRLQVNKLQ 900
901 SPRGFRISAK ISPKLEKIEF SIHCQEIAGL GKTIVYSTDH LASQFSAKTP NIRKDLNSLE 960
961 KCLFYESSSD GEVGKTSRVS HKDGLEESLS SDDSILDYLN ERKKSSKAVE SAAVIHPEAH1020
1021 SSSHFSNGRQ VRSNGNYECF HSCKDKTQCR HLCCKEGIPV KYIKEKGPSS IKPVSKPDQI1080
1081 RQPLLAKNIN TTPHLEKRLN SKPKQWQEEN TDIATVHTLP SKIYNLSQQM SSMEAGEQVL1140
1141 KSGPENCPEI IPIDLESSDS YSSNTAASSI SDPNGDLDFL GSDIEFE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
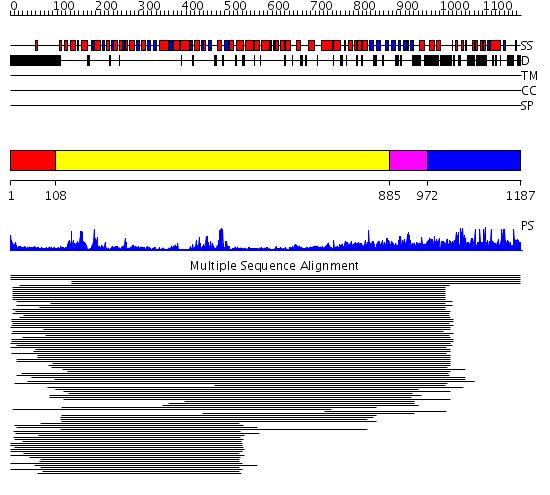
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 37.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 2 | View Details | [108..884] | 6.522879 | Helicase-like "domain" of reverse gyrase; Topoisomerase "domain" of reverse gyrase | |
| 3 | View Details | [885..971] | 3.065994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [972..1187] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)