













| General Information: |
|
| Name(s) found: |
SNT2 /
YGL131C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1403 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
DNA binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKEEDFQLP RRREAAKNVN YNEMEIDTKL VQQIQIAEKS GAKTKGSNSQ TPRNCKRTSN 60
61 PASRNEKFKY QKFLHDKNTC WNFIPTLPPS FRKNSRFSNI LDLDDAMIDL KKMSLFNTES 120
121 VLLSANDTIY MISEPAGEPY YVGRVVNFVS KPEFSNTIHE AIKTTSVFPA KFFQVRMNWF 180
181 YRPRDIQEHV NTFNPRLVYA SLHQDICPIS SYRGKCSIFH KDEVFDVLPN EKECIIRPNI 240
241 FYFDELFDRY TLKYYKVYST DKILNKWNSK SPFLYVLNRR FRYIYTEPKY PLENVLKKYV 300
301 FHELEVNELS PADYQWDKRC QFCKEWCIQK ESLSCDECGV CAHLYCMDPP LDRKPNKDVV 360
361 WTCFSCLQKQ QGTKDSHVRF LEEQALELDF IRSVRQKIEE ISSKAIKENV GYNTENCWFQ 420
421 YLGIYSISHI GDALNDSMFF PYPFKPSRVG VKYQWNGCNH NVPWRRNSYI SANSEEERGS 480
481 TKTSELAWVL DASKITTRKL SEYIEQCKSE ICPILNVRGE TCNFIDVVLK NLLFTNYDTA 540
541 EAFKKCKREL SRKFLKEPSF TAVEIRKFEE AVEKFGSELR PVCEYVGTQP MSMIVRFYYN 600
601 WKKTERGLTV RGKLSKLSKN KRKKEIANHE NDVETKYIDD SSFDTEKLSL AESSFQCMFC 660
661 KTDYSPMWYR VTGGSDDEKI KIRMQTGVNE KTEISEKSPA HSKKNEKLGA LCIRCARMWR 720
721 RYAIKWVPPL ETLRKITGTC QNSFYSAIEG IIEENNTNKF TLSPFQAHNK LLEWELVQDS 780
781 ELIIRQRMKV YKNPNSFVKM KRYSMTFHTQ LYKMAVRSYR KNEFHPETMQ RDLELFIEDN 840
841 KEVRKAIPEQ KPERAKNTKD EFPVNIIRQS PGTIKTSDTS RNRKCNDVFI EKASNNNIPK 900
901 ITNASNDLIE ISIKTGGSSS GSVSVDKGFK FVKFDNKTFQ RLRNSLKLVN NKLPKYNEPS 960
961 TKKIKMINDI ALSNPLNEPN GASYNYTVIS HSKETSVALE KYHDGNKPSK MLEKDMILKH1020
1021 TKNKPKNPDT AWANNSARTF CSVCKEKFND NDNYEVVCGN CGLTVHYFCY AIKLPKDMKK1080
1081 NTNLKTFKWL CDPCSNDLNP IISTTYQCSM CPTKDYDYDR YRSQSFKICP DALKCTSLGT1140
1141 WVHLVCSLFN EDIKYGNGQS MQPALNTTAV LIKHSRFTCG VCRINGGGLV KCNKCQYRYH1200
1201 ITCAQNSSNF KLMFEKKNMS VDTTLPCIKD VKLNDTYTLR PILICDRHDI SLEGNELYPL1260
1261 SYKPQHTLSY IEQYCRYYKC ESDHSLVELR YFEQLRLRHG EMPGNSHDSA IKPKIYVLPF1320
1321 ERTCPHCGTN KSLYWYEDII CHSCNLRSGA QELDFDSASA NISNDNGLPV EITQQLMEGI1380
1381 EPAMFDIDIS EAGTDKNTHP SSQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
SNT2 XBP1 |
| View Details | Gavin AC, et al. (2002) |
|
ADH1 ENO2 FAR11 FAR3 FBA1 GPM1 HFM1 HSC82 PDC1 ROM2 SNT2 SSE2 TPI1 |
| View Details | Ho Y, et al. (2002) |
|
SNT2 YLR413W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
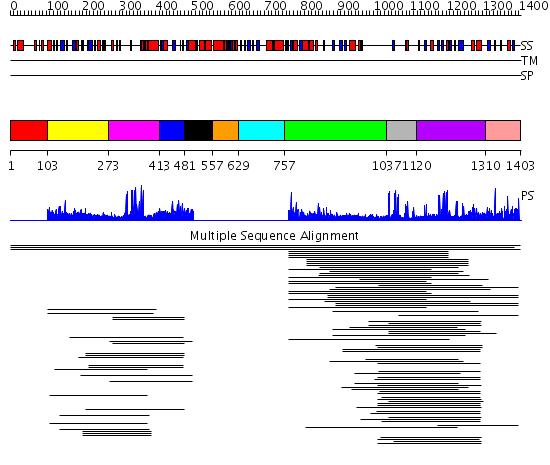
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [103..272] | 31.420216 | BAH domain No confident structure predictions are available. | |
| 3 | View Details | [273..412] | 7.045757 | Nuclear corepressor KAP-1 (TIF-1beta) | |
| 4 | View Details | [413..480] | 1.040987 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [481..556] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [557..628] | 10.376751 | Myb-like DNA-binding domain No confident structure predictions are available. | |
| 7 | View Details | [629..756] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [757..1036] | 5.218996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1037..1119] | 11.853872 | PHD-finger No confident structure predictions are available. | |
| 10 | View Details | [1120..1309] | 6.247989 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1310..1403] | 3.60206 | GATA zinc finger No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)