













| General Information: |
|
| Name(s) found: |
XBP1 /
YIL101C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
response to stress
[IDA |
| Molecular Function: |
transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKYPAFSINS DTVHLTDNPL DDYQRLYLVS VLDRDSPPAS FSAGLNIRKV NYKSSIAAQF 60
61 THPNFIISAR DAGNGEEAAA QNVLNCFEYQ FPNLQTIQSL VHEQTLLSQL ASSATPHSAL 120
121 HLHDKNILMG KIILPSRSNK TPVSASPTKQ EKKALSTASR ENATSSLTKN QQFKLTKMDH 180
181 NLINDKLINP NNCVIWSHDS GYVFMTGIWR LYQDVMKGLI NLPRGDSVST SQQQFFCKAE 240
241 FEKILSFCFY NHSSFTSEES SSVLLSSSTS SPPKRRTSTG STFLDANASS SSTSSTQANN 300
301 YIDFHWNNIK PELRDLICQS YKDFLINELG PDQIDLPNLN PANFTKRIRG GYIKIQGTWL 360
361 PMEISRLLCL RFCFPIRYFL VPIFGPDFPK DCESWYLAHQ NVTFASSTTG AGAATAATAA 420
421 ANTSTNFTST AVARPRQKPR PRPRQRSTSM SHSKAQKLVI EDALPSFDSF VENLGLSSND 480
481 KNFIKKNSKR QKSSTYTSQT SSPIGPRDPT VQILSNLASF YNTHGHRYSY PGNIYIPQQR 540
541 YSLPPPNQLS SPQRQLNYTY DHIHPVPSQY QSPRHYNVPS SPIAPAPPTF PQPYGDDHYH 600
601 FLKYASEVYK QQNQRPAHNT NTNMDTSFSP RANNSLNNFK FKTNSKQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
SNT2 XBP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
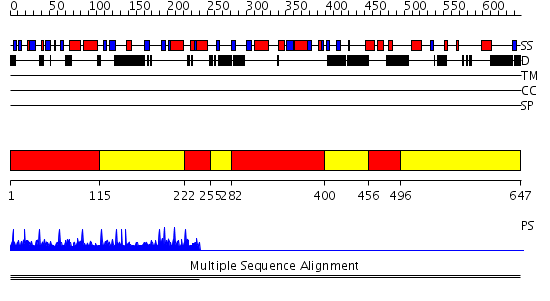
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] [222..254] [282..399] [456..495] |
7.03 | Adenovirus hexon | |
| 2 | View Details | [115..221] [255..281] [400..455] [496..647] |
7.03 | Adenovirus hexon | |
| 3 | View Details | [442..647] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)