













| General Information: |
|
| Name(s) found: |
RBG1 /
YAL036C
[SGD]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 369 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
ribosome biogenesis
[RCA |
| Molecular Function: |
GTP binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTTVEKIKA IEDEMARTQK NKATSFHLGQ LKAKLAKLRR ELLTSASSGS GGGAGIGFDV 60
61 ARTGVASVGF VGFPSVGKST LLSKLTGTES EAAEYEFTTL VTVPGVIRYK GAKIQMLDLP 120
121 GIIDGAKDGR GRGKQVIAVA RTCNLLFIIL DVNKPLHHKQ IIEKELEGVG IRLNKTPPDI 180
181 LIKKKEKGGI SITNTVPLTH LGNDEIRAVM SEYRINSAEI AFRCDATVDD LIDVLEASSR 240
241 RYMPAIYVLN KIDSLSIEEL ELLYRIPNAV PISSGQDWNL DELLQVMWDR LNLVRIYTKP 300
301 KGQIPDFTDP VVLRSDRCSV KDFCNQIHKS LVDDFRNALV YGSSVKHQPQ YVGLSHILED 360
361 EDVVTILKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
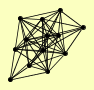
|
CBF5 FUN12 KRE33 MAK5 RBG1 RPL15A RPS13 RPS1B RPS4B, RPS4A SBP1 SRO9 STM1 YGR054W |
| View Details | Krogan NJ, et al. (2006) |
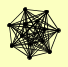
|
HFM1 LYS4 NEW1 RBG1 SGD1 SOK1 SSZ1 TCB1 TGL5 TMA46 YEF3 YLR072W ZUO1 |
| View Details | Gavin AC, et al. (2006) |

|
RBG1 TIF4631 |
| View Details | Gavin AC, et al. (2002) |

|
ASC1 CBF5 CDC33 DBP2 HEK2 HHF1, HHF2 LHP1 MIS1 MRP7 NAB6 POL2 PUS7 RBG1 ROM2 SBP1 SNP1 SRO9 STM1 STO1 SUI3 TIF1, TIF2 TIF4631 TIF4632 UTP20 YMR310C |
| View Details | Ho Y, et al. (2002) |
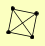
|
CLU1 RBG1 RPN1 TIF1, TIF2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
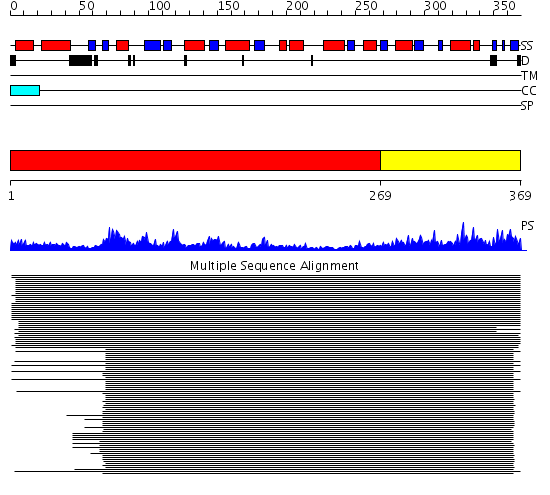
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..268] | 33.69897 | GTPase Era, N-terminal domain; GTPase Era C-terminal domain | |
| 2 | View Details | [269..369] | 1.348996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)