













| General Information: |
|
| Name(s) found: |
DBP2 /
YNL112W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 546 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA nuclear outer membrane [IMP |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay
[IPI |
| Molecular Function: |
RNA helicase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTYGGRDQQY NKTNYKSRGG DFRGGRNSDR NSYNDRPQGG NYRGGFGGRS NYNQPQELIK 60
61 PNWDEELPKL PTFEKNFYVE HESVRDRSDS EIAQFRKENE MTISGHDIPK PITTFDEAGF 120
121 PDYVLNEVKA EGFDKPTGIQ CQGWPMALSG RDMVGIAATG SGKTLSYCLP GIVHINAQPL 180
181 LAPGDGPIVL VLAPTRELAV QIQTECSKFG HSSRIRNTCV YGGVPKSQQI RDLSRGSEIV 240
241 IATPGRLIDM LEIGKTNLKR VTYLVLDEAD RMLDMGFEPQ IRKIVDQIRP DRQTLMWSAT 300
301 WPKEVKQLAA DYLNDPIQVQ VGSLELSASH NITQIVEVVS DFEKRDRLNK YLETASQDNE 360
361 YKTLIFASTK RMCDDITKYL REDGWPALAI HGDKDQRERD WVLQEFRNGR SPIMVATDVA 420
421 ARGIDVKGIN YVINYDMPGN IEDYVHRIGR TGRAGATGTA ISFFTEQNKG LGAKLISIMR 480
481 EANQNIPPEL LKYDRRSYGG GHPRYGGGRG GRGGYGRRGG YGGGRGGYGG NRQRDGGWGN 540
541 RGRSNY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
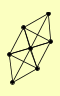
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
DBP2 FUN12 MIS1 RPL15A RPL27B RPP0 STM1 |
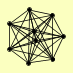
| View Details | Krogan NJ, et al. (2006) |
|
CWP2 DBP2 FLC2 PPN1 RPS31 SRP40 TFA1 TFA2 YAP1801 |
| View Details | Gavin AC, et al. (2006) |

|
DBP2 GYL1 RPL2A, RPL2B |
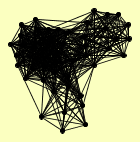
| View Details | Gavin AC, et al. (2002) |
|
ADE3 ARX1 ASC1 CBF5 CDC33 CKA1 CKA2 CKB1 DBP2 HEK2 HHF1, HHF2 LHP1 LSG1 MIS1 MRP7 MRPS28 NAB6 NOG1 NOP7 NUG1 NUP120 NUP145 NUP84 NUP85 POL2 PUS7 RAT1 RBG1 ROM2 RPP2B SBP1 SEC13 SEC16 SEC23 SEC31 SEH1 SHM1 SNP1 SRO9 STM1 STO1 SUI3 TIF1, TIF2 TIF4631 TIF4632 UTP20 YMR310C YPL009C |
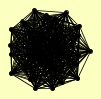
| View Details | Ho Y, et al. (2002) |
|
CKA1 CKB1 CKB2 DBP2 EGD1 ENP2 ERB1 FPR3 HAS1 HHT1, HHT2 HOT1 HTA1 HTB2 KRE33 KRI1 MGM101 NOG1 NOP12 NOP2 NOP4 NOP56 NUG1 PDI1 POB3 PUF6 PWP1 SPT16 SSF1 TIF6 YER084W |
| View Details | Qiu et al. (2008) |
|
DBP2 DBP3 NOP13 POP2 RAT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
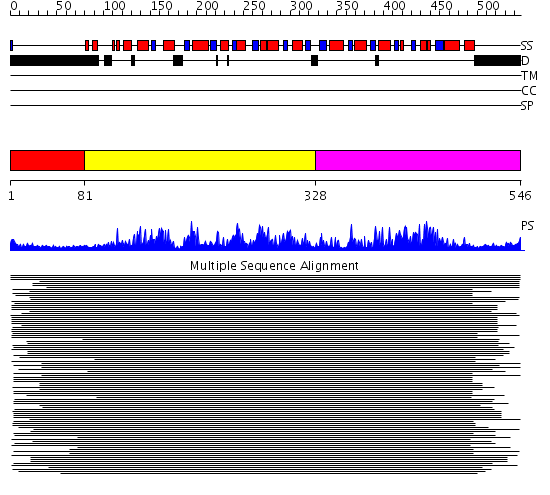
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..327] | 730.0103 | Putative DEAD box RNA helicase | |
| 3 | View Details | [328..546] | 730.0103 | Putative DEAD box RNA helicase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)