













| General Information: |
|
| Name(s) found: |
ACS2 /
YLR153C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
acetyl-CoA biosynthetic process
[IDA histone acetylation [IMP |
| Molecular Function: |
acetate-CoA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIKEHKVVY EAHNVKALKA PQHFYNSQPG KGYVTDMQHY QEMYQQSINE PEKFFDKMAK 60
61 EYLHWDAPYT KVQSGSLNNG DVAWFLNGKL NASYNCVDRH AFANPDKPAL IYEADDESDN 120
121 KIITFGELLR KVSQIAGVLK SWGVKKGDTV AIYLPMIPEA VIAMLAVARI GAIHSVVFAG 180
181 FSAGSLKDRV VDANSKVVIT CDEGKRGGKT INTKKIVDEG LNGVDLVSRI LVFQRTGTEG 240
241 IPMKAGRDYW WHEEAAKQRT YLPPVSCDAE DPLFLLYTSG STGSPKGVVH TTGGYLLGAA 300
301 LTTRYVFDIH PEDVLFTAGD VGWITGHTYA LYGPLTLGTA SIIFESTPAY PDYGRYWRII 360
361 QRHKATHFYV APTALRLIKR VGEAEIAKYD TSSLRVLGSV GEPISPDLWE WYHEKVGNKN 420
421 CVICDTMWQT ESGSHLIAPL AGAVPTKPGS ATVPFFGINA CIIDPVTGVE LEGNDVEGVL 480
481 AVKSPWPSMA RSVWNHHDRY MDTYLKPYPG HYFTGDGAGR DHDGYYWIRG RVDDVVNVSG 540
541 HRLSTSEIEA SISNHENVSE AAVVGIPDEL TGQTVVAYVS LKDGYLQNNA TEGDAEHITP 600
601 DNLRRELILQ VRGEIGPFAS PKTIILVRDL PRTRSGKIMR RVLRKVASNE AEQLGDLTTL 660
661 ANPEVVPAII SAVENQFFSQ KKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ACS2 TKL1 TKL2 |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACS2 ADE3 ADE6 ADH4 ADO1 ALA1 APE2 ARA1 ARG4 BAT1 CAR2 CDC10 CDC60 CLU1 CPR1 CYS3 DED81 DLD3 DPS1 DUG1 ENP1 ERG10 ERG20 FUM1 GCY1 GPH1 GRS1 HOM2 HYP2 ILS1 ILV3 IMD3 KRS1 LSP1 MDH3 MPC54 NAS6 OLA1 PAB1 PDC5 PFK2 PGM2 PMI40 PRP6 RHO1 SAC6 SCP160 SOD1 SSZ1 SUI1 THR4 TPS1 TRR1 UBA1 WTM1 YDL124W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
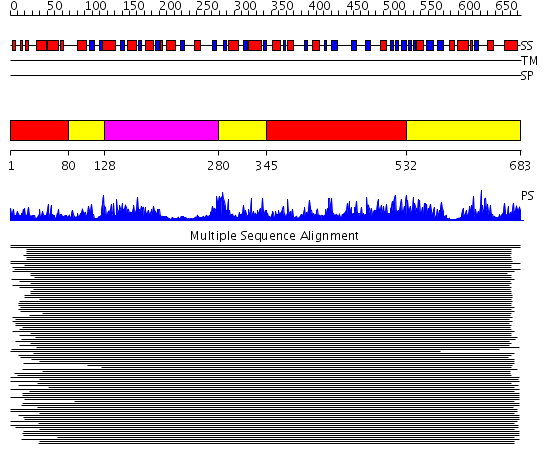
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] [345..531] |
269.218487 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 2 | View Details | [80..127] [280..344] [532..683] |
269.218487 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 3 | View Details | [128..279] | 269.218487 | Phenylalanine activating domain of gramicidin synthetase 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)