













| General Information: |
|
| Name(s) found: |
ILV3 /
YJR016C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 585 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
branched chain family amino acid biosynthetic process
[TAS |
| Molecular Function: |
dihydroxy-acid dehydratase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLLTKVATS RQFSTTRCVA KKLNKYSYII TEPKGQGASQ AMLYATGFKK EDFKKPQVGV 60
61 GSCWWSGNPC NMHLLDLNNR CSQSIEKAGL KAMQFNTIGV SDGISMGTKG MRYSLQSREI 120
121 IADSFETIMM AQHYDANIAI PSCDKNMPGV MMAMGRHNRP SIMVYGGTIL PGHPTCGSSK 180
181 ISKNIDIVSA FQSYGEYISK QFTEEEREDV VEHACPGPGS CGGMYTANTM ASAAEVLGLT 240
241 IPNSSSFPAV SKEKLAECDN IGEYIKKTME LGILPRDILT KEAFENAITY VVATGGSTNA 300
301 VLHLVAVAHS AGVKLSPDDF QRISDTTPLI GDFKPSGKYV MADLINVGGT QSVIKYLYEN 360
361 NMLHGNTMTV TGDTLAERAK KAPSLPEGQE IIKPLSHPIK ANGHLQILYG SLAPGGAVGK 420
421 ITGKEGTYFK GRARVFEEEG AFIEALERGE IKKGEKTVVV IRYEGPRGAP GMPEMLKPSS 480
481 ALMGYGLGKD VALLTDGRFS GGSHGFLIGH IVPEAAEGGP IGLVRDGDEI IIDADNNKID 540
541 LLVSDKEMAQ RKQSWVAPPP RYTRGTLSKY AKLVSNASNG CVLDA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
ACS2 CAR2 DLD3 DUG1 ERG10 FUM1 ILV3 RHO1 WTM1 |
| View Details | Qiu et al. (2008) |

|
ILV1 ILV3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
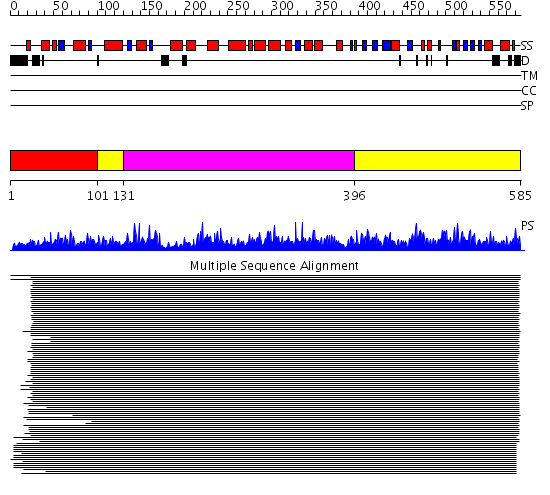
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 9.48 | Histidine ammonia-lyase (HAL) | |
| 2 | View Details | [101..130] [396..585] |
8.9 | Putative glycerate kinase (hypothetical protein TM1585) | |
| 3 | View Details | [131..395] | 8.9 | Putative glycerate kinase (hypothetical protein TM1585) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)