













| General Information: |
|
| Name(s) found: |
BRE2 /
YLR015W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 505 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Set1C/COMPASS complex
[IPI |
| Biological Process: |
transcription
[ISS histone methylation [IDA telomere maintenance [IMP chromatin silencing at telomere [IMP |
| Molecular Function: |
histone methyltransferase activity (H3-K4 specific)
[IDA transcription regulator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLGIIPYQE GTDIVYKNAL QGQQEGKRPN LPQMEATHQI KSSVQGTSYE FVRTEDIPLN 60
61 RRHFVYRPCS ANPFFTILGY GCTEYPFDHS GMSVMDRSEG LSISRDGNDL VSVPDQYGWR 120
121 TARSDVCIKE GMTYWEVEVI RGGNKKFADG VNNKENADDS VDEVQSGIYE KMHKQVNDTP 180
181 HLRFGVCRRE ASLEAPVGFD VYGYGIRDIS LESIHEGKLN CVLENGSPLK EGDKIGFLLS 240
241 LPSIHTQIKQ AKEFTKRRIF ALNSHMDTMN EPWREDAENG PSRKKLKQET TNKEFQRALL 300
301 EDIEYNDVVR DQIAIRYKNQ LFFEATDYVK TTKPEYYSSD KRERQDYYQL EDSYLAIFQN 360
361 GKYLGKAFEN LKPLLPPFSE LQYNEKFYLG YWQHGEARDE SNDKNTTSAK KKKQQQKKKK 420
421 GLILRNKYVN NNKLGYYPTI SCFNGGTARI ISEEDKLEYL DQIRSAYCVD GNSKVNTLDT 480
481 LYKEQIAEDI VWDIIDELEQ IALQQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
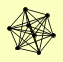
|
BRE2 SDC1 SET1 SHG1 SPP1 SWD1 SWD2 SWD3 |
| View Details | Krogan NJ, et al. (2006) |
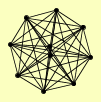
|
BRE2 ERF2 SDC1 SET1 SHG1 SPP1 SWD1 SWD3 YHI9 YLL023C |
| View Details | Gavin AC, et al. (2006) |
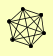
|
BRE2 SDC1 SET1 SPP1 SWD1 SWD3 |
| View Details | Gavin AC, et al. (2002) |
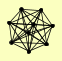
|
BRE2 DOA4 RSM7 SDC1 SET1 SPP1 SSZ1 SWD1 SWD3 |
| View Details | Ho Y, et al. (2002) |

|
BRE2 SET1 |
| View Details | Qiu et al. (2008) |
|
BRE2 SDC1 SET1 SHG1 SPP1 SWD1 SWD2 SWD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
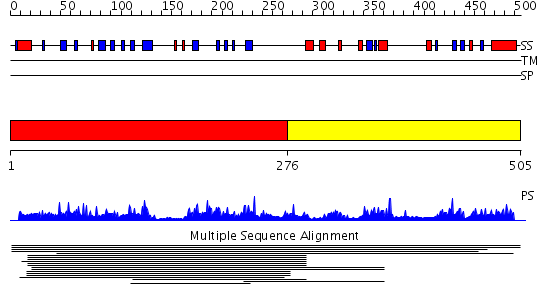
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..275] | 3.009939 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [276..505] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [293..505] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)