













| General Information: |
|
| Name(s) found: |
UBR2 /
YLR024C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1872 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
protein polyubiquitination
[TAS protein monoubiquitination [TAS |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDSDLSITN IRDFLTELPK LAKCEYSETT SYLLWKTLNL RLKHSDNDIN WRSLVSILNS 60
61 EAWENEKYRD ILNGRKWRTL EFENDHHSVG NMHIGTACTR LCFPSETIYY CFTCSTNPLY 120
121 EICELCFDKE KHVNHSYVAK VVMRPEGRIC HCGDPFAFND PSDAFKCKNE LNNIPISNDN 180
181 SNVTDDENVI SLLNYVLDFL IDVTVSYKEE AEAHSSERKA SSLMHPNQNS ITDDIMEKHE 240
241 CEPLVNDENF VFFDNNWSNT RKEAHMEWAI QIEEEECNVH YMDLASTITR ILNTPVEYAI 300
301 SITKALEDSH DVVTVLQSEN FFEIDQIAKE FQKENIVVHV RKADDIFKRK LTDDLTDWLY 360
361 SLCFKAATSL QNKYALRISM LDVWYSHFSK MRVSPTNTNP DFSKINLLGG FLISNEDSDE 420
421 SWFKPWSLEN IEDERISKIL TNYNERLIRA HSPNTVSHFY NFYGSRFQYI IINSINILSK 480
481 KSKFKMLKIM ASLFSLRDES RKFLAAQYID VYLSVLYDAV ASDAKECQVT LMSILGQYTF 540
541 QDPSIANMTI SSGFIERTIR FAFTLMAFNP EDLMSYLPIS LYNGFKLPTE TIRNRRTIIC 600
601 FKDLCTIMSA NTVPEELLSN EAIFNAIIES FSEFSNVLPL KRETKEHVEV ENFDFSAFYF 660
661 FFSSILIMTD GYTRSISLVK DAAFRKQIVL KLLDVAQTRE FESLTNSRKA ISPDNASTNE 720
721 NDSNKATLST VRETICNYVA ETINFQVGVN TQYFFNPMSY LFKFVIQWSQ CGRYEPIPAS 780
781 LTNYINLFEV FQDKQKALYI SESALSTLVL IGQINVGFWV RNGTPITHQA RMYTKYSMRE 840
841 FTYISDIFNV QFSMAMCNPD ELMVTYLSRW GLKHWANGVP MYDYPDTETT VAVVNECILL 900
901 LIQLLTEVRS LVMKSSKEGF ERTFKSEIIH ALCFDTCSYA QIVNCIPEHI TKHPSFDIYL 960
961 EKYANYTSPV SLTDNGIFVL KEKYKDEIDP YYIGLSSSRR YDVEKNIRLN MANLKKMKYE1020
1021 DTFVPAKKVK DLLKNTLFSG LYSISSVNTF GLFLKNTLDH IIKYDYDNLL PRVVHLIHLC1080
1081 VVNNLNEFMG ILWHEYAIVD TEFCHYHSIG SILYYCLLKD NFSESHGKIR EIFRYLMETA1140
1141 PHVNVNSYLR EQTTSYTPGI LWPTKEDKSH KDKEFERKKH LARLRKKKLM KKLAQQQMKF1200
1201 MENNSVDTSD ISTPRTTSPS LSPTRINAEN SSNTINSCCD DDCVFCKMPK DDDVFVYFSY1260
1261 QERNICDHGI DFTNPTDVNR INSLFSGKQT KDSAIQENPQ DDDGTRLKFT SCEPVLRACG1320
1321 HGSHTKCLSG HMKSIRGIQN QTTKNIPLSY GSGLIYCPVC NSLSNSFLPK TNDIDKRTSS1380
1381 QFFMCIEKRS EAEENLDPMS SICIKAAMIL GDLQGKKVTT IEDAYKVVNS VFINTISNTE1440
1441 LRLRSHKKEG KIVNMERISS QCILTLHLVC ELKSFIYKKF VNSKTFSSEI SRKIWNWNEF1500
1501 LIKGNNVNLL LYMSQNFDNI DGGKTPQPPN LCIYEMFKRR FHQLLLLLAR DMMRVNFYKD1560
1561 CRNKIKISSN GSEEPSTSFS YLFNTFKKYV DLFKPDDVRF DFTSLEKIKD FICSLLLESL1620
1621 SIFCRRTFLL FNIQYDDDGD GDNNNNRSNN FMDVKQREIE LIFRYFKLPN LTHFLKDFFY1680
1681 NELTQNIERY NDGNDNLRIQ QVIYDMVQNI NTRAYPSPEH IQLIELPLNL SKFSLDNDEI1740
1741 SNKCDKYEIA VCLLCGQKCH IQKSIALQGY LQGECTDHMR NGCEITSAYG VFLMTGTNAI1800
1801 YLSYGKRGTF YAAPYLSKYG ETNEDYKFGT PVYLNRARYA NLANEIVFGN MIPHIVFRLT1860
1861 DGSADLGGWE TM |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
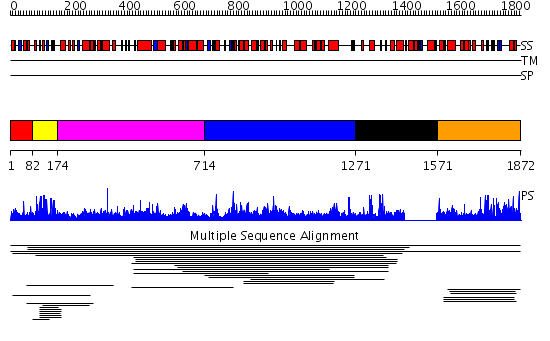
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [82..173] | 32.958607 | Putative zinc finger in N-recognin No confident structure predictions are available. | |
| 3 | View Details | [174..713] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [714..1270] | 2.015884 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1271..1570] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1571..1872] | 5.006938 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1017..1169] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1170..1375] | 1.054979 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1376..1576] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1577..1714] | 1.033991 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1715..1872] | 3.038988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)