













| General Information: |
|
| Name(s) found: |
PHO23 /
YNL097C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 330 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA histone deacetylase complex [IDA |
| Biological Process: |
chromatin modification
[IMP histone deacetylation [IDA |
| Molecular Function: |
histone deacetylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSPANLFPG LNDITDVLEE FPLATSRYLT LLHEIDAKCV HSMPNLNERI DKFLKKDFNK 60
61 DHQTQVRLLN NINKIYEELM PSLEEKMHVS SIMLDNLDRL TSRLELAYEV AIKNTEIPRG 120
121 LRLGVDNHPA MHLHHELMEK IESKSNSKSS QALKSESRRE AMAANRRQGE HYSASTHQQD 180
181 DSKNDANYGG SRHESQDHTG NNTNSRKRAN AANTNNADPE TKKRKRRVAT TAVSPSTIST 240
241 ATAVNNGRIG TSTASRGVSS VGNSNNSRIS RPKTNDYGEP LYCYCNQVAY GEMVGCDGAD 300
301 CELEWFHLPC IGLETLPKGK WYCDDCKKKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CTI6 DEP1 EAF3 PHO23 RCO1 RPD3 RXT2 RXT3 SAP30 SDS3 SIN3 UME1 |
| View Details | Krogan NJ, et al. (2006) |
|
ASH1 COQ6 CTI6 DEP1 EAF3 GRX5 PHO23 RCO1 RPD3 RXT2 RXT3 SAP30 SAY1 SDS3 SIN3 UME1 |
| View Details | Gavin AC, et al. (2002) |

|
ABF2 CKA1 CKA2 CKB1 CKB2 CTI6 DEP1 DOT6 EAF3 ECM16 HTA2 IOC3 ISW1 MED4 NHP10 NRD1 PHO23 RCO1 RPD3 RSC8 RXT2 RXT3 SAP30 SDS3 SIN3 STO1 TOP1 TUP1 UME1 UME6 VPS1 |
| View Details | Qiu et al. (2008) |
|
HDA1 HDA3 HOS1 HST1 PHO23 RPD3 RXT2 SAP30 SDS3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
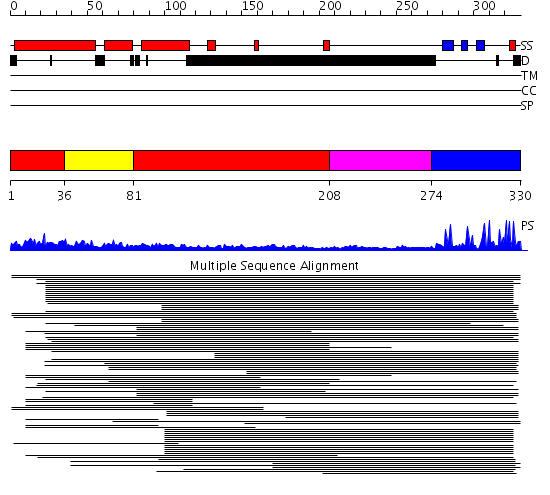
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..35] [81..207] |
7.33 | Variant surface glycoprotein (N-terminal domain) | |
| 2 | View Details | [36..80] | 7.33 | Variant surface glycoprotein (N-terminal domain) | |
| 3 | View Details | [208..273] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [274..330] | 5.69897 | Williams-Beuren syndrome transcription factor, WSTF |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)