













| General Information: |
|
| Name(s) found: |
UME6 /
YDR207C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 836 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
ascospore formation
[IMP positive regulation of meiosis [IMP reciprocal meiotic recombination [IMP response to drug [IMP negative regulation of transcription, mitotic [IMP histone deacetylation [IDA |
| Molecular Function: |
DNA binding
[IDA histone deacetylase activity [IDA transcription regulator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDKARSQSK HMDESNAAAS LLSMETTANN HHYLHNKTSR ATLMNSSQDG KKHAEDEVSD 60
61 GANSRHPTIS SASIESLKTT YDENPLLSIM KSTCAPNNTP VHTPSGSPSL KVQSGGDIKD 120
121 DPKENDTTTT TNTTLQDRRD SDNAVHAAAS PLAPSNTPSD PKSLCNGHVA QATDPQISGA 180
181 IQPQYTATNE DVFPYSSTST NSNTATTTIV AGAKKKIHLP PPQAPAVSSP GTTAAGSGAG 240
241 TGSGIRSRTG SDLPLIITSA NKNNGKTTNS PMSILSRNNS TNNNDNNSIQ SSDSRESSNN 300
301 NEIGGYLRGG TKRGGSPSND SQVQHNVHDD QCAVGVAPRN FYFNKDREIT DPNVKLDENE 360
361 SKINISFWLN SKYRDEAYSL NESSSNNASS NTDTPTNSRH ANTSSSITSR NNFQHFRFNQ 420
421 IPSQPPTSAS SFTSTNNNNP QRNNINRGED PFATSSRPST GFFYGDLPNR NNRNSPFHTN 480
481 EQYIPPPPPK YINSKLDGLR SRLLLGPNSA SSSTKLDDDL GTAAAVLSNM RSSPYRTHDK 540
541 PISNVNDMNN TNALGVPASR PHSSSFPSKG VLRPILLRIH NSEQQPIFES NNSTAVFDED 600
601 QDQNQDLSPY HLNLNSKKVL DPTFESRTRQ VTWNKNGKRI DRRLSAPEQQ QQLEVPPLKK 660
661 SRRSVGNARV ASQTNSDYNS LGESSTSSAP SSPSLKASSG LAYTADYPNA TSPDFAKSKG 720
721 KNVKPKAKSK AKQSSKKRPN NTTSKSKANN SQESNNATSS TSQGTRSRTG CWICRLRKKK 780
781 CTEERPHCFN CERLKLDCHY DAFKPDFVSD PKKKQMKLEE IKKKTKEAKR RAMKKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
SAP30 SDS3 UME6 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
IME1 UME6 |
| View Details | Krogan NJ, et al. (2006) |
|
ALG5 BRE5 CSN12 CUP2 DBP7 LUG1 MET10 MSC7 MSE1 NOP53 NPY1 PBP4 RPF2 RRS1 SAS5 UME6 YGR126W YHR127W YPR045C YRA1 |
| View Details | Gavin AC, et al. (2002) |

|
ABF2 CKA1 CKA2 CKB1 CKB2 CTI6 DEP1 DOT6 EAF3 ECM16 HTA2 IOC3 ISW1 MED4 NHP10 NRD1 PHO23 RCO1 RPD3 RSC8 RXT2 RXT3 SAP30 SDS3 SIN3 STO1 TOP1 TUP1 UME1 UME6 VPS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
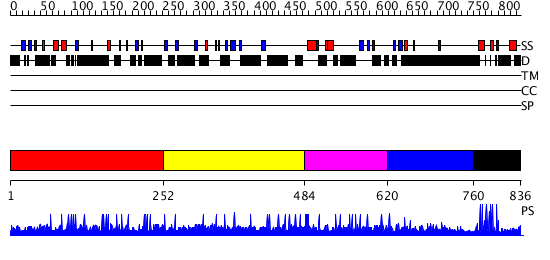
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [252..483] | 14.37 | No description for 1je5A was found. | |
| 3 | View Details | [484..619] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [620..759] | 3.123997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [760..836] | 57.917458 | Hap1 (Cyp1); HAP1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)