













| General Information: |
|
| Name(s) found: |
BRE5 /
YNR051C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
protein deubiquitination
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVTVQDICF AFLQNYYERM RTDPSKLAYF YASTAELTHT NYQSKSTNEK DDVLPTVKVT 60
61 GRENINKFFS RNDAKVRSLK LKLDTIDFQY TGHLHKSILI MATGEMFWTG TPVYKFCQTF 120
121 ILLPSSNGST FDITNDIIRF ISNSFKPYVL TDASLSQSNE ENSVSAVEED KIRHESGVEK 180
181 EKEKEKSPEI SKPKAKKETV KDTTAPTESS TQEKPIVDHS QPRAIPVTKE SKIHTETVPS 240
241 STKGNHKQDE VSTEELGNVT KLNEKSHKAE KKAAPIKTKE GSVEAINAVN NSSLPNGKEV 300
301 SDEKPVPGGV KEAETEIKPI EPQVSDAKES GNNASTPSSS PEPVANPPKM TWASKLMNEN 360
361 SDRISKNNTT VEYIRPETLP KKPTERKFEM GNRRDNASAN SKNKKKPVFS TVNKDGFYPI 420
421 YIRGTNGLRE EKLRSALEKE FGKVMRITAA DNFAVVDFET QKSQIDALEK KKKSIDGIEV 480
481 CLERKTVKKP TSNNPPGIFT NGTRSHRKQP LKRKD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
BRE5 FIT1 UBP3 ZEO1 |
| View Details | Krogan NJ, et al. (2006) |

|
ALG5 BRE5 CSN12 CUP2 DBP7 LUG1 MET10 MSC7 MSE1 NOP53 NPY1 PBP4 RPF2 RRS1 SAS5 UME6 YGR126W YHR127W YPR045C YRA1 |
| View Details | Gavin AC, et al. (2006) |

|
BRE5 UBP3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
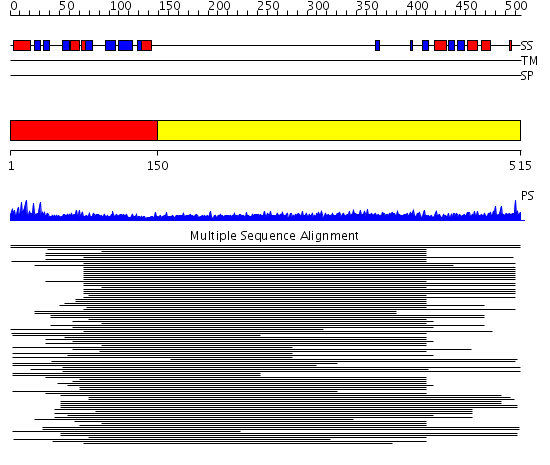
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 9.522879 | Nuclear transport factor-2 (NTF2) | |
| 2 | View Details | [150..515] | 14.63 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)