













| General Information: |
|
| Name(s) found: |
MSC7 /
YHR039C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 644 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA |
| Biological Process: |
reciprocal meiotic recombination
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVYLNSDM INHLNSTVQA YFNLWLEKQN AIMRSQPQII QDNQKLIGIT TLVASIFTLY 60
61 VLVKIISTPA KCSSSYKPVK FSLPAPEAAQ NNWKGKRSVS TNIWNPEEPN FIQCHCPATG 120
121 QYLGSFPSKT EADIDEMVSK AGKAQSTWGN SDFSRRLRVL ASLHDYILNN QDLIARVACR 180
181 DSGKTMLDAS MGEILVTLEK IQWTIKHGQR ALQPSRRPGP TNFFMKWYKG AEIRYEPLGV 240
241 ISSIVSWNYP FHNLLGPIIA ALFTGNAIVV KCSEQVVWSS EFFVELIRKC LEACDEDPDL 300
301 VQLCYCLPPT ENDDSANYFT SHPGFKHITF IGSQPVAHYI LKCAAKSLTP VVVELGGKDA 360
361 FIVLDSAKNL DALSSIIMRG TFQSSGQNCI GIERVIVSKE NYDDLVKILN DRMTANPLRQ 420
421 GSDIDHLENV DMGAMISDNR FDELEALVKD AVAKGARLLQ GGSRFKHPKY PQGHYFQPTL 480
481 LVDVTPEMKI AQNEVFGPIL VMMKAKNTDH CVQLANSAPF GLGGSVFGAD IKECNYVANS 540
541 LQTGNVAIND FATFYVCQLP FGGINGSGYG KFGGEEGLLG LCNAKSVCFD TLPFVSTQIP 600
601 KPLDYPIRNN AKAWNFVKSF IVGAYTNSTW QRIKSLFSLA KEAS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
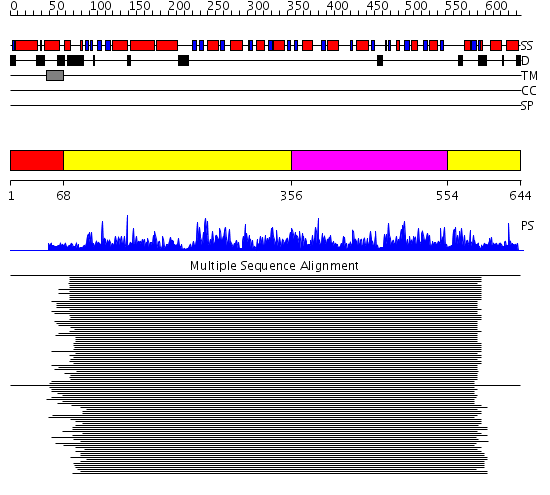
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [68..355] [554..644] |
1476.9897 | Aldehyde reductase (dehydrogenase), ALDH | |
| 3 | View Details | [356..553] | 1476.9897 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)