













| General Information: |
|
| Name(s) found: |
PSO2 /
YMR137C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 661 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
DNA repair
[IMP double-strand break repair [TAS |
| Molecular Function: |
5'-3' exonuclease activity
[IDA damaged DNA binding [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKSIVQIR RSEVKRKRSS TASSTSEGKT LHKNTHTSSK RQRTLTEFNI PTSSNLPVRS 60
61 SSYSFSRFSC STSNKNTEPV IINDDDHNSI CLEDTAKVEI TIDTDEEELV SLHDNEVSAI 120
121 ENRTEDRIVT ELEEQVNVKV STEVIQCPIC LENLSHLELY ERETHCDTCI GSDPSNMGTP 180
181 KKNIRSFISN PSSPAKTKRD IATSKKPTRV KLVLPSFKII KFNNGHEIVV DGFNYKASET 240
241 ISQYFLSHFH SDHYIGLKKS WNNPDENPIK KTLYCSKITA ILVNLKFKIP MDEIQILPMN 300
301 KRFWITDTIS VVTLDANHCP GAIIMLFQEF LANSYDKPIR QILHTGDFRS NAKMIETIQK 360
361 WLAETANETI DQVYLDTTYM TMGYNFPSQH SVCETVADFT LRLIKHGKNK TFGDSQRNLF 420
421 HFQRKKTLTT HRYRVLFLVG TYTIGKEKLA IKICEFLKTK LFVMPNSVKF SMMLTVLQNN 480
481 ENQNDMWDES LLTSNLHESS VHLVPIRVLK SQETIEAYLK SLKELETDYV KDIEDVVGFI 540
541 PTGWSHNFGL KYQKKNDDDE NEMSGNTEYC LELMKNDRDN DDENGFEISS ILRQYKKYNK 600
601 FQVFNVPYSE HSSFNDLVKF GCKLKCSEVI PTVNLNNLWK VRYMTNWFQC WENVRKTRAA 660
661 K |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
AIR1 AIR2 MTR4 PAP2 PSO2 RPL18A, RPL18B STE4 TRF5 |
| View Details | Ho Y, et al. (2002) |
|
MGM101 PSO2 PTC7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
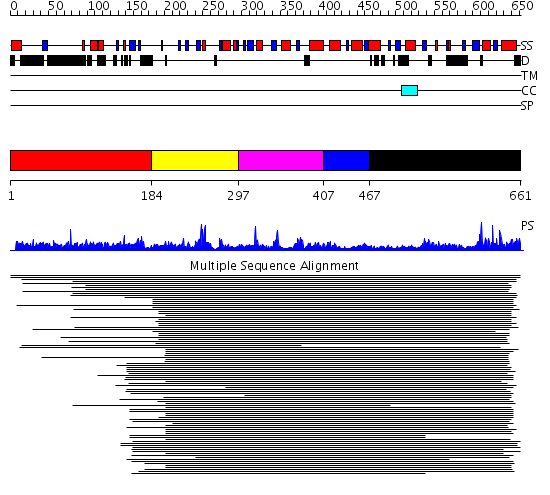
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [184..296] | 16.30103 | Zn metallo-beta-lactamase | |
| 3 | View Details | [297..406] | 16.30103 | Zn metallo-beta-lactamase | |
| 4 | View Details | [407..466] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [467..661] | 1.15397 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)