













| General Information: |
|
| Name(s) found: |
UBI4 /
YLL039C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 381 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC |
| Biological Process: |
protein deubiquitination
[TAS ascospore formation [TAS protein polyubiquitination [TAS protein monoubiquitination [TAS response to stress [TAS protein ubiquitination [IMP |
| Molecular Function: |
protein tag
[ISS ATP-dependent protein binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQIFVKTLTG KTITLEVESS DTIDNVKSKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN 60
61 IQKESTLHLV LRLRGGMQIF VKTLTGKTIT LEVESSDTID NVKSKIQDKE GIPPDQQRLI 120
121 FAGKQLEDGR TLSDYNIQKE STLHLVLRLR GGMQIFVKTL TGKTITLEVE SSDTIDNVKS 180
181 KIQDKEGIPP DQQRLIFAGK QLEDGRTLSD YNIQKESTLH LVLRLRGGMQ IFVKTLTGKT 240
241 ITLEVESSDT IDNVKSKIQD KEGIPPDQQR LIFAGKQLED GRTLSDYNIQ KESTLHLVLR 300
301 LRGGMQIFVK TLTGKTITLE VESSDTIDNV KSKIQDKEGI PPDQQRLIFA GKQLEDGRTL 360
361 SDYNIQKEST LHLVLRLRGG N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
ADH2 ADR1 AHP1 ARF1 ARP2 ARP7 ATP3 BBC1 BEM2 BFR2 BIR1 BNA5 CAR2 CDC16 CDC33 CDC39 CDC53 CDC7 COF1 COP1 CPR1 CRM1 DIG1 DIG2 DMA1 DMA2 DSK2 DUF1 DUN1 DUR1,2 ECM10 ECM29 ECM33 EDE1 ELC1 ERB1 ERG1 FAA4 FAR1 FET4 FOL2 FPR3 GAL3 GAL83 GCN1 GFA1 GIS4 GPD1 GPD2 GRR1 GUF1 GUS1 GYP5 HAS1 HHF1, HHF2 HMF1 HOM6 HRT1 HRT3 HTB2 HXT6 HYP2 IDH1 JIP3 KRE33 KRI1 KSS1 LHS1 LOC1 LSP1 MCM4 MDM30 MDN1 MES1 MET30 MET4 MKT1 MMS1 MMS4 MSE1 MUS81 MYO2 NAP1 NHP2 NIP1 NOP12 NOP2 NPL4 NUT1 PDC5 PDC6 PFK1 PHO84 PIB1 PIL1 PIM1 PMA1 PMA2 POR1 PRB1 PST2 PWP1 PYC1 RAD1 RAD23 RAD53 RAD7 RGC1 RNR2 RPA135 RPA190 RPC10 RPC19 RPF2 RPG1 RPN1 RPN10 RPT2 RPT4 RVB1 RVS167 SAH1 SAN1 SAR1 SAW1 SEC23 SEC27 SEC6 SEN1 SES1 SHP1 SKP1 SNF1 SNF4 SRP1 SRV2 STE12 STE20 STE7 STI1 SWD1 TAF14 TEC1 THI3 TIF34 TIF35 TPS1 TSL1 UBC7 UBI4 UBP7 UFD2 UTP22 VAC14 VPS13 YAR009C YDR049W YDR239C YGP1 YHB1 YHR033W YLR035C-A YNL311C YPL113C YRA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | experimental1 - mih1-1 | McCusker D, et al (2007) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #09 Alpha Factor Prep4-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
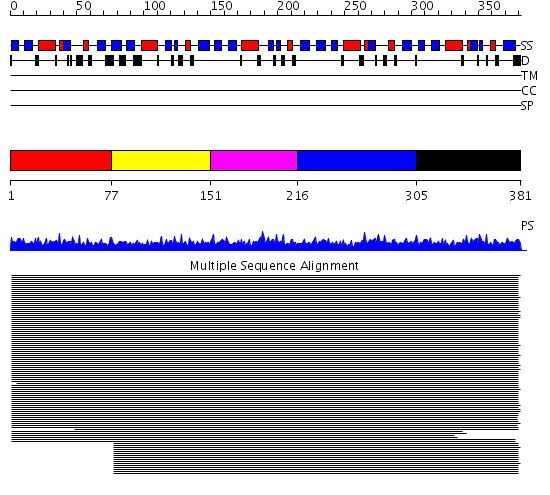
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 181.68867 | Nedd8 | |
| 2 | View Details | [77..150] | 14.91 | Nedd8 | |
| 3 | View Details | [151..215] | 193.37734 | Ubiquitin | |
| 4 | View Details | [216..304] | 337.07631 | Ubiquitin | |
| 5 | View Details | [305..381] | 13.19 | Ubiquitin-like N-terminal domain of PLIC-2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)